Nomenclature
Short Name:
LIMK1
Full Name:
LIM domain kinase 1
Alias:
- EC 2.7.11.1
- Kinase LIMK1
- KIZ
- KIZ-1
- LIMK
- LIMK-1
Classification
Type:
Protein-serine/threonine kinase
Group:
TKL
Family:
LISK
SubFamily:
LIMK
Specific Links
Structure
Mol. Mass (Da):
72585
# Amino Acids:
647
# mRNA Isoforms:
4
mRNA Isoforms:
72,585 Da (647 AA; P53667); 70,793 Da (633 AA; P53667-2); 68,729 Da (613 AA; P53667-4); 33,373 Da (305 AA; P53667-3)
4D Structure:
Self-associates. The LIM domain interacts with the cytoplasmic domain of NRG1. Binds ROCK1. Interacts with SSH1. Interacts with NISCH By similarity. Interacts with RLIM and RNF6 By similarity. Interacts with TPPP.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
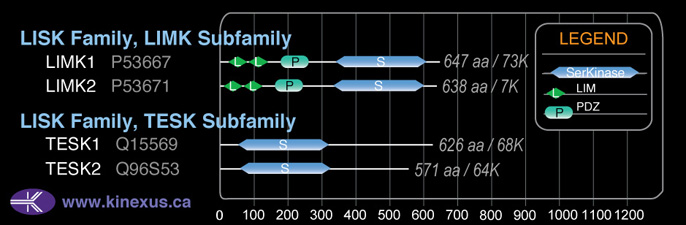
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S210, S274, S286, S287, S296, S298, S302, S307, S310, S313, S323+, S337+, S497+, S534, S637.
Threonine phosphorylated:
T229, T253, T270, T508+.
Tyrosine phosphorylated:
Y415.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 37
37
645
45
1068
 2
2
42
18
50
 4
4
71
18
94
 23
23
398
138
684
 26
26
450
32
354
 1
1
17
113
77
 9
9
156
43
464
 65
65
1140
59
2188
 14
14
252
24
180
 4
4
73
112
144
 2
2
41
40
62
 34
34
592
234
548
 4
4
65
51
114
 2
2
36
12
29
 5
5
81
34
99
 0.7
0.7
13
22
9
 2
2
30
131
50
 7
7
114
28
111
 2
2
40
129
72
 15
15
267
162
313
 9
9
155
29
149
 5
5
95
35
126
 4
4
71
29
75
 6
6
99
27
125
 4
4
71
29
90
 100
100
1745
84
3623
 4
4
65
54
87
 5
5
91
27
83
 4
4
72
27
75
 8
8
142
42
136
 12
12
213
30
255
 33
33
582
51
543
 18
18
316
88
597
 42
42
728
83
629
 2
2
43
48
35
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 98.6
98.6
99.2
100 -
-
-
96 -
-
-
98 -
-
-
- 94.1
94.1
96.3
94 -
-
-
- 95.2
95.2
96.9
95 95.2
95.2
96.9
95 -
-
-
- 53.5
53.5
69.4
- 82
82
87
84.5 72.2
72.2
81.1
77 73
73
82.3
74 -
-
-
- 26.8
26.8
35.7
51 34.5
34.5
43.9
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PAK1 - Q13153 |
| 2 | YWHAZ - P63104 |
| 3 | PXN - P49023 |
| 4 | SSH3 - Q8TE77 |
| 5 | CFL2 - Q9Y281 |
| 6 | PAK2 - Q13177 |
| 7 | LATS1 - O95835 |
| 8 | MAPK14 - Q16539 |
| 9 | RLIM - Q9NVW2 |
| 10 | ROCK2 - O75116 |
| 11 | BRAF - P15056 |
| 12 | SSH1 - Q8WYL5 |
Regulation
Activation:
Phosphorylation at Ser-323 and Thr-508 increases phosphotransferase activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
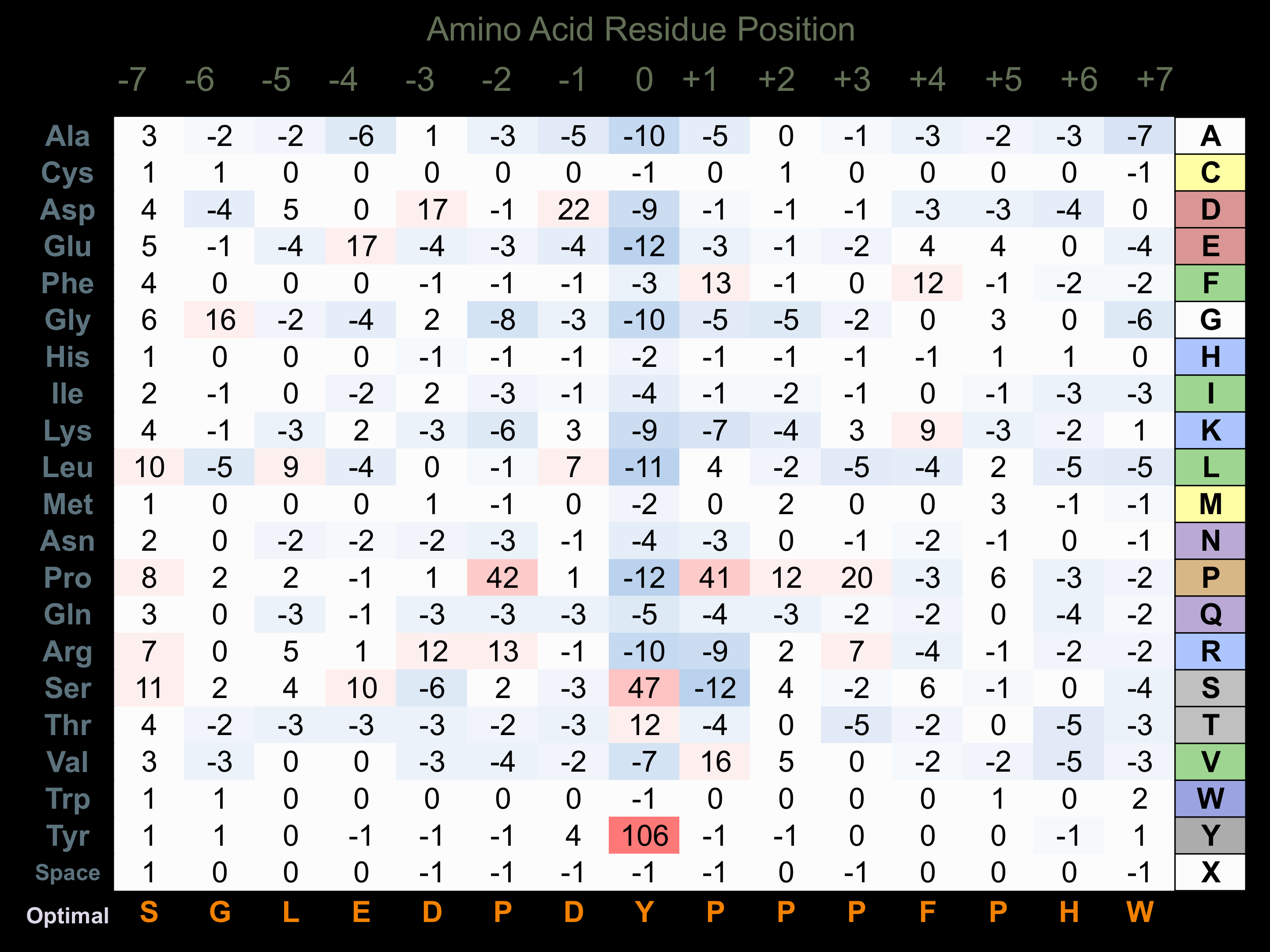
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Developmental, and cardiovascular disorders
Specific Diseases (Non-cancerous):
Williams syndrome (WS); Intracranial aneurysm; Williams-Beuren syndrome
Comments:
Williams Syndrome (WS) is characterized by slight cognitive impairment, learning difficulties, highly anxious but empathetic character, abnormal facial characteristics (short nose, broad nose tip, puffy eye sockets, wide mouth, small chin, full lips and cheeks, long neck, short stature, sloping shoulders, limited mobility, a high risk of developing diabetes in their 30's, and cardiovascular issues. Intracranial Aneurysm is characterized by the ballooning, or bulging of an artery in the brain leading to a brain aneurysm. Williams-Beuren Syndrome is a rare condition with a relation to the supravalvular aortic stenosis and Williams Syndrome disorders. Williams-Beuren Syndrome can affect kidney, bone, or skin tissues.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= +87, p<0.039); and Gastric cancer (%CFC= +63, p<0.001). The COSMIC website notes an up-regulated expression score for LIMK1 in diverse human cancers of 669, which is 1.5-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 27 for this protein kinase in human cancers was 0.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. The mutations C84S, G177E + L178A, and T508EE can enhance aggregation of actin, whereas D460N, and R503G + K504A + K505A can fully inhibit actin aggregation, and T508V/E can partially inhibit actin aggregation. Activation of LIMK1 by ROCK1 can be fully inhibited with a T508A mutation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25034 diverse cancer specimens. This rate is only -4 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.35 % in 805 skin cancers tested; 0.31 % in 1093 large intestine cancers tested; 0.29 % in 589 stomach cancers tested; 0.26 % in 602 endometrium cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,197 cancer specimens
Comments:
Only 2 deletions, 2 insertions, and no complex mutations are noted on the COSMIC website.

