Nomenclature
Short Name:
MEK4
Full Name:
Dual specificity mitogen-activated protein kinase kinase 4
Alias:
- C-Jun N- terminal kinase kinase 1
- C-JUN N-terminal kinase kinase 1
- Kinase SEK1
- MAP2K4
- SEK1
- SERK1
- EC 2.7.12.2
- JNK activating kinase 1
- JNK kinase 1
- JNKK1
Classification
Type:
Dual specificity protein kinase
Group:
STE
Family:
STE7
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
44,288
# Amino Acids:
399
# mRNA Isoforms:
2
mRNA Isoforms:
45,584 Da (410 AA; P45985-2); 44,288 Da (399 AA; P45985)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 102 | 367 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K131.
Methylated:
R58.
Serine phosphorylated:
S5, S7, S18, S26, S80-, S90, S257+, S393, S394.
Threonine phosphorylated:
T56, T78+, T261+.
Tyrosine phosphorylated:
Y269-, Y397.
Ubiquitinated:
K140, K143.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
962
29
1046
 6
6
58
20
36
 8
8
76
12
30
 56
56
538
102
522
 83
83
799
28
736
 7
7
69
75
41
 47
47
452
42
606
 38
38
370
48
526
 62
62
601
20
576
 9
9
91
109
46
 7
7
70
38
44
 53
53
513
231
624
 6
6
61
34
25
 5
5
47
20
39
 6
6
56
31
34
 9
9
90
17
61
 5
5
45
294
51
 7
7
67
25
43
 14
14
133
112
97
 71
71
684
112
709
 12
12
118
33
79
 7
7
69
35
59
 6
6
54
30
39
 6
6
57
27
34
 7
7
64
32
46
 54
54
523
62
664
 8
8
79
40
35
 6
6
55
26
38
 5
5
46
27
36
 19
19
183
28
99
 83
83
802
24
712
 59
59
565
36
582
 26
26
254
70
370
 86
86
827
62
758
 26
26
252
35
190
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 35.3
35.3
53.6
99.5 97.3
97.3
97.3
100 -
-
-
100 -
-
-
- 89.3
89.3
90.6
99 -
-
-
- 98
98
98.3
98 41.8
41.8
57.3
98 -
-
-
- -
-
-
- 36.6
36.6
53.9
96.5 77.3
77.3
80.9
93 48.4
48.4
64.4
91 -
-
-
- 54
54
65.8
64 56
56
68.8
- 44.6
44.6
61.6
54 55.9
55.9
71.2
- -
-
-
- -
-
-
- -
-
-
- 31.1
31.1
48.4
- 28.3
28.3
38.9
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | MAPK8 - P45983 |
| 2 | EFNA3 - P52797 |
| 3 | EFNA4 - P52798 |
| 4 | MAPK8IP3 - Q9UPT6 |
| 5 | MAP3K2 - Q9Y2U5 |
| 6 | MAPK14 - Q16539 |
| 7 | MAPK10 - P53779 |
| 8 | MAP3K10 - Q02779 |
| 9 | MAP3K8 - P41279 |
| 10 | MAP3K7 - O43318 |
| 11 | AKT1 - P31749 |
| 12 | MAP3K14 - Q99558 |
| 13 | MAPK9 - P45984 |
| 14 | MAPK12 - P53778 |
| 15 | MAP3K11 - Q16584 |
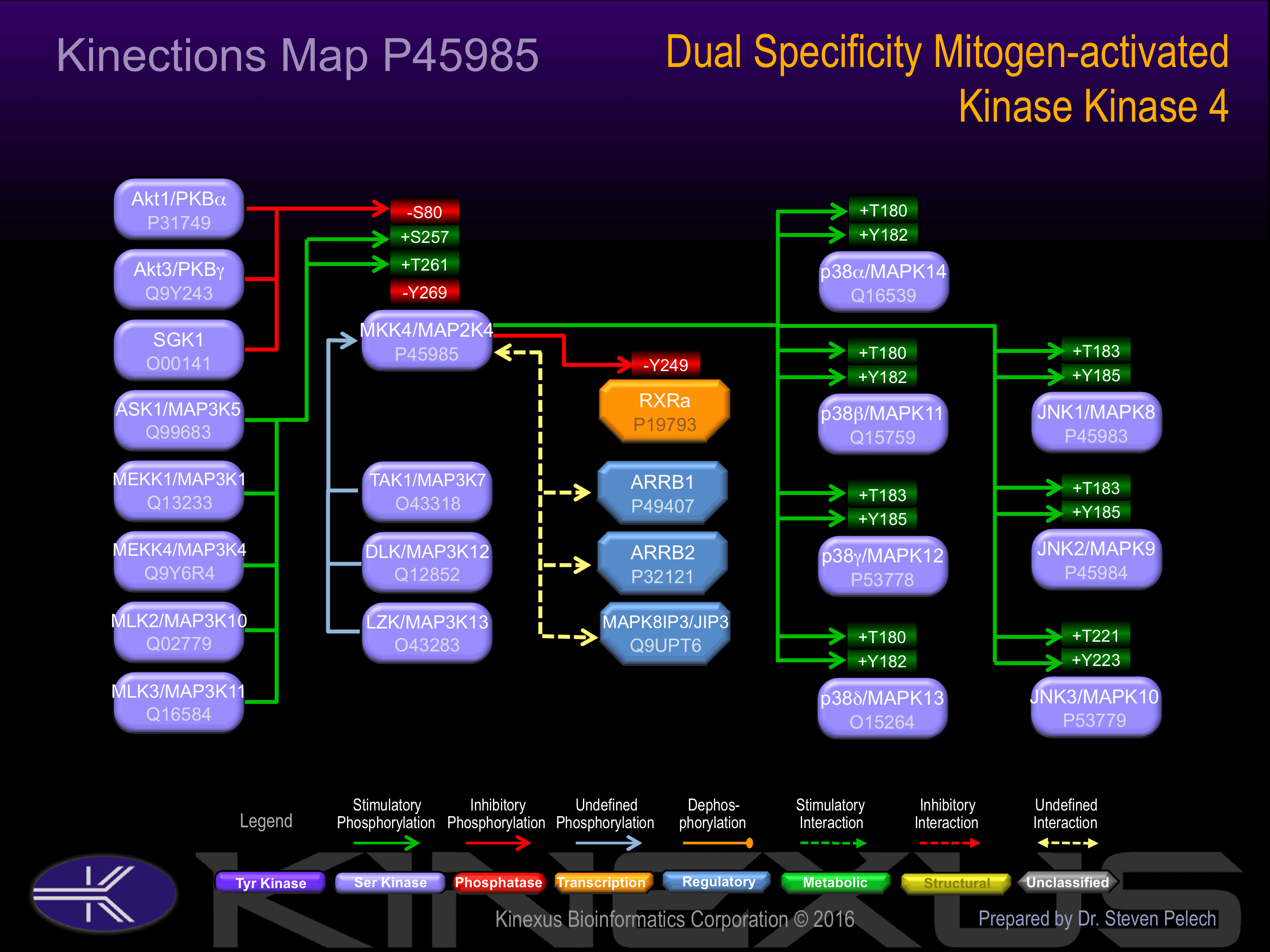
Regulation
Activation:
Phosphorylation of Ser-257 and Thr-261 increases phosphotransferase activity.
Inhibition:
Phosphorylation of Ser-80 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| SGK | O00141 | S80 | IERLRTHSIESSGKL | - |
| Akt1 | P31749 | S80 | IERLRTHSIESSGKL | - |
| AKT3 | Q9Y243 | S80 | IERLRTHSIESSGKL | - |
| MEKK4 | Q9Y6R4 | S257 | ISGQLVDSIAKTRDA | + |
| MEKK1 | Q13233 | S257 | ISGQLVDSIAKTRDA | + |
| ASK1 | Q99683 | S257 | ISGQLVDSIAKTRDA | + |
| MLK2 | Q02779 | S257 | ISGQLVDSIAKTRDA | + |
| MLK3 | Q16584 | S257 | ISGQLVDSIAKTRDA | + |
| MEKK4 | Q9Y6R4 | T261 | LVDSIAKTRDAGCRP | + |
| MEKK1 | Q13233 | T261 | LVDSIAKTRDAGCRP | + |
| ASK1 | Q99683 | T261 | LVDSIAKTRDAGCRP | + |
| MLK2 | Q02779 | T261 | LVDSIAKTRDAGCRP | + |
| MLK3 | Q16584 | T261 | LVDSIAKTRDAGCRP | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| JNK1 (MAPK8) | P45983 | T183 | AGTSFMMTPYVVTRY | + |
| JNK1 (MAPK8) | P45983 | Y185 | TSFMMTPYVVTRYYR | + |
| JNK2 (MAPK9) | P45984 | T183 | ACTNFMMTPYVVTRY | + |
| JNK2 (MAPK9) | P45984 | Y185 | TNFMMTPYVVTRYYR | + |
| JNK3 (MAPK10) | P53779 | T221 | AGTSFMMTPYVVTRY | + |
| JNK3 (MAPK10) | P53779 | Y223 | TSFMMTPYVVTRYYR | + |
| p38a MAPK (MAPK14) | Q16539 | T180 | RHTDDEMTGYVATRW | + |
| p38a MAPK (MAPK14) | Q16539 | Y182 | TDDEMTGYVATRWYR | + |
| p38b MAPK (MAPK11) | Q15759 | T180 | RQADEEMTGYVATRW | + |
| p38b MAPK (MAPK11) | Q15759 | Y182 | ADEEMTGYVATRWYR | + |
| p38d MAPK (MAPK13) | O15264 | T180 | RHADAEMTGYVVTRW | + |
| p38d MAPK (MAPK13) | O15264 | Y182 | ADAEMTGYVVTRWYR | + |
| p38g MAPK (MAPK12) | P53778 | T183 | RQADSEMTGYVVTRW | + |
| p38g MAPK (MAPK12) | P53778 | Y185 | ADSEMTGYVVTRWYR | + |
| RXRa | P19793 | Y249 | VEPKTETYVEANMGL | - |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
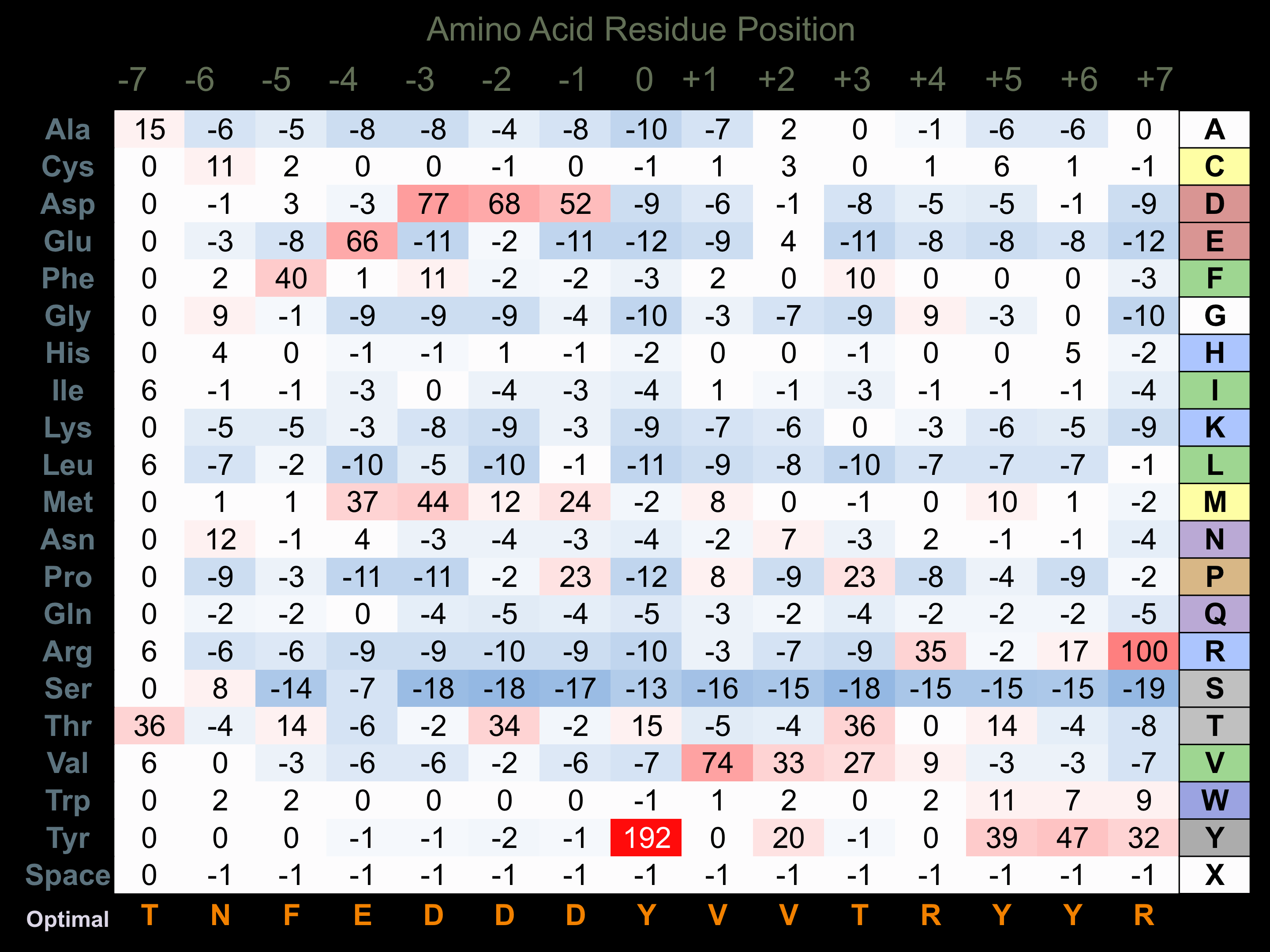
Matrix Type:
Experimentally derived from alignment of 15 known protein substrate phosphosites and 18 peptides phosphorylated by recombinant MEK4 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, neurological disorders
Specific Diseases (Non-cancerous):
Alzheimer's disease (AD)
Comments:
The plasma level of MEK4 protein was found to negatively correlate with earlier detection of Alzheimer's Disease in a twin study.
Specific Cancer Types:
Pancreatic cancer; Prostate cancer
Comments:
MEK4 appears to be a tumour suppressor protein (TSP), but it may also promote metastasis. Cancer-related mutations in human tumours point to a loss of function of the protein kinase. The active form of the protein kinase normally acts to inhibit tumour cell proliferation. Significantly elevated expression of MEK4 has been observed in invasive pancreatic cancer specimens, indicating a possible role for the protein in tumorigenesis. In animal studies, mice injected with pancreatic cancer cells over-expressing MEK4 displayed increased occurence of metastasis, implicating the MEK4 protein as a key promotor of metastasis in pancreatic cancer cells. In addition, the over-expression of MEK4 protein did not effect either cell growth or migration. MEK4 over-expression is correlated with increased expression of heat shock protein 27 (HSP27) and matrix metalloproteinase 2 (MMP-2) protein, which are required for the increased invasive phenotype of the cells. Therefore, MEK4 appears to have an important role in the promotion of metastasis in pancreatic cancer cells.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= -46, p<0.006); Bladder carcinomas (%CFC= +49, p<(0.0003); Cervical cancer (%CFC= +101, p<0.0001); Cervical cancer stage 1B (%CFC= +113); Cervical cancer stage 2A (%CFC= +231); and Cervical cancer stage 2B (%CFC= +470). The COSMIC website notes an up-regulated expression score for MEK4 in diverse human cancers of 297, which is 0.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 680 for this protein kinase in human cancers was 11.3-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.21 % in 28157 diverse cancer specimens. This rate is 2.8-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 1.02 % in 1712 large intestine cancers tested; 0.55 % in 2498 breast cancers tested; 0.46 % in 603 endometrium cancers tested; 0.43 % in 1338 pancreas cancers tested; 0.34 % in 811 skin cancers tested; 0.19 % in 2023 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R134W (9); S184L (6).
Comments:
Eleven deletions, 7 insertions and no complex mutations are noted on the COSMIC website. Although there are many mutations observed in cancer specimens for MEK4, there are very few mutations in the first 70 amino acid residues in the kinase.