Nomenclature
Short Name:
ATM
Full Name:
Serine-protein kinase ATM
Alias:
- A-T, mutated
- ATD
- ATDC
- Kinase ATM
- TEL1
- TELO1; telomere maintenance 1
- ATA
- Ataxia telangiectasia mutated
- Ataxia telangiectasia mutated homolog
- ATC
Classification
Type:
Protein-serine/threonine kinase
Group:
Atypical
Family:
PIKK
SubFamily:
ATM
Specific Links
Structure
Mol. Mass (Da):
350,687
# Amino Acids:
3056
# mRNA Isoforms:
1
mRNA Isoforms:
350,687 Da (3056 AA; Q13315)
4D Structure:
Dimers or tetramers in inactive state. On DNA damage, autophosphorylation dissociates ATM into monomers rendering them catalytically active. Binds p53/TP53, ABL1, BRCA1, NBN/nibrin and TERF1. Part of the BRCA1-associated genome surveillance complex (BASC), which contains BRCA1, MSH2, MSH6, MLH1, ATM, BLM, PMS2 and the RAD50-MRE11-NBN protein complex. This association could be a dynamic process changing throughout the cell cycle and within subnuclear domains. Interacts with RAD17; DNA damage promotes the association. Interacts with EEF1E1; the interaction, induced on DNA damage, upregulates TP53. Interacts with DCLRE1C, MYST1, KAT5, OBFC2B, ATMIN and CEP164. Interacts with AP2B1 and AP3B2; the interaction occurs in cytoplasmic vesicles.Interacts with TELO2 AND TTI1.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
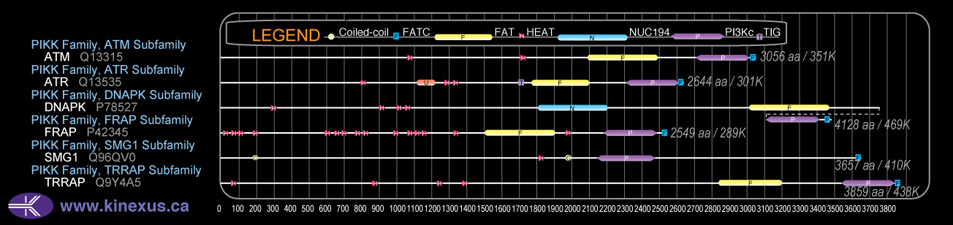
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K3016 (N6).
Methylated:
K1126, K1656, K2643.
Serine phosphorylated:
S47, S49, S83, S85, S200, S214, S305, S310, S367+, S381, S440, S569, S644, S646, S794+, S821, S1403, S1655, S1673, S1878, S1883, S1891, S1893, S1974, S1981+, S2162, S2165, S2168, S2310, S2592, S2996+.
Threonine phosphorylated:
T72, T86, T202, T237, T297, T373, T935, T1662, T1885, T1908, T1985, T1997, T2031, T2751.
Tyrosine phosphorylated:
Y54, Y303, Y313, Y332, Y370, Y380, Y980, Y1717, Y1753, Y1763, Y1915, Y2019, Y2170, Y2969.
Ubiquitinated:
K1101, K1109, K1114, K1572, K2025, K2148.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 45
45
1026
72
1074
 2
2
43
26
39
 3
3
75
66
56
 12
12
282
265
697
 29
29
661
58
540
 5
5
123
148
305
 8
8
187
84
390
 21
21
469
114
1523
 23
23
521
27
469
 3
3
67
192
59
 4
4
100
100
107
 23
23
530
340
594
 17
17
392
99
393
 3
3
79
21
75
 6
6
137
91
145
 3
3
68
38
75
 6
6
143
444
1499
 10
10
218
80
194
 3
3
58
195
45
 23
23
529
234
502
 5
5
122
90
159
 14
14
323
96
331
 6
6
139
93
160
 3
3
72
81
63
 9
9
209
91
219
 25
25
555
186
1000
 12
12
274
105
254
 4
4
87
82
75
 4
4
101
81
92
 4
4
89
42
54
 41
41
935
42
632
 100
100
2261
63
7539
 10
10
231
125
613
 33
33
743
171
694
 4
4
85
87
69
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.4
99.4
99.7
99 -
-
-
98 -
-
-
90 -
-
-
- -
-
-
90 -
-
-
- 83.9
83.9
91.3
84 -
-
-
84 -
-
-
- -
-
-
- 69.6
69.6
82.8
70 20.6
20.6
40
- 35.1
35.1
46.4
58 -
-
-
- 22.1
22.1
42.9
27 -
-
-
- -
-
-
28 36.3
36.3
56.8
- -
-
-
- -
-
-
- -
-
-
- 23.9
23.9
42.5
38 -
-
-
- 20.8
20.8
40.9
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Phosphorylation at Ser-1981 increases phosphotransferase activity and induces interaction with ATM, NBS1 and p53.
Inhibition:
NA
Synthesis:
By ionizing radiation.
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| 4E-BP1 | Q13541 | S93 | DEPPMEASQSHLRNS | |
| 53BP1 | Q12888 | S25 | PCLIIEDSQPESQVL | |
| 53BP1 | Q12888 | S29 | IEDSQPESQVLEDDS | |
| 53BP1 | Q12888 | S6 | __MDPTGSQLDSDFS | |
| 53BP1 | Q12888 | S784 | GVEKCSDSQSWEDIA | |
| Abl1 | P00519 | S446 | PYPGIDLSQVYELLE | + |
| Abl1 iso2 | P00519-2 | S465 | PYPGIDLSQVYELLE | + |
| Akt1 (PKBa) | P31749 | S473 | RPHFPQFSYSASGTA | + |
| APLF | Q8IW19 | S116 | MQCTLRNSQVLDEDN | |
| ATF2 | P15336 | S490 | QSTEPALSQIVMAPS | |
| ATF2 | P15336 | S498 | QIVMAPSSQSQPSGS | |
| ATM | Q13315 | S1893 | PANLDSESEHFFRCC | |
| ATM | Q13315 | S1981 | SLAFEEGSQSTTISS | + |
| ATM | Q13315 | S367 | DTRSLEISQSYTTTQ | + |
| ATM | Q13315 | S440 | SPLLMILSQLLPQQR | |
| BLM | P54132 | T99 | NAPAGQETQRGGSKS | |
| BRCA1 | P38398 | S1189 | QKGELSRSPSPFTHT | |
| BRCA1 | P38398 | S1387 | EDCSGLSSQSDILTT | |
| BRCA1 | P38398 | S1423 | AVLEQHGSQPSNSYP | |
| BRCA1 | P38398 | S1457 | SEKAVLTSQKSSEYP | |
| BRCA1 | P38398 | S1497 | EPGVERSSPSKCPSL | |
| BRCA1 | P38398 | S1524 | LQNRNYPSQEELIKV | |
| BRCA1 | P38398 | S1542 | EEQQLEESGPHDLTE | |
| Chk1 (CHEK1) | O14757 | S317 | ENVKYSSSQPEPRTG | + |
| Chk1 (CHEK1) | O14757 | S345 | LVQGISFSQPTCPDH | + |
| Chk2 (CHEK2) | O96017 | S19 | SHGSSACSQPHGSVT | + |
| Chk2 (CHEK2) | O96017 | S28 | PHGSVTQSQGSSSQS | + |
| Chk2 (CHEK2) | O96017 | S33 | TQSQGSSSQSQGISS | + |
| Chk2 (CHEK2) | O96017 | S35 | SQGSSSQSQGISSSS | + |
| Chk2 (CHEK2) | O96017 | S50 | TSTMPNSSQSSHSSS | |
| Chk2 (CHEK2) | O96017 | T26 | SQPHGSVTQSQGSSS | + |
| Chk2 (CHEK2) | O96017 | T68 | SSLETVSTQELYSIP | + |
| CREB1 | P16220 | S111 | TIAESEDSQESVDSV | - |
| CREB1 | P16220 | S121 | SVDSVTDSQKRREIL | |
| CREB1 | P16220 | T100 | LKRLFSGTQISTIAE | |
| Ctip | Q99708 | S664 | IDPGADLSQYKMDVT | |
| Ctip | Q99708 | S745 | SCLADSFSQAADEEE | |
| DNAPK (PRKDC) | P78527 | S2612 | MFVETQASQGTLQTR | ? |
| DNAPK (PRKDC) | P78527 | T2609 | LTPMFVETQASQGTL | + |
| DNAPK (PRKDC) | P78527 | T2638 | VAGQIRATQQQHDFT | - |
| DNAPK (PRKDC) | P78527 | T2647 | QQHDFTLTQTADGRS | - |
| DYRK2 | Q92630 | S442 | IELLGMPSQKLLDAS | + |
| DYRK2 | Q92630 | T106 | NKRTVLTTQPNGLTT | + |
| E2F1 | Q01094 | S31 | ALRLLDSSQIVIISA | |
| FANCD2 | Q9BXW9 | S222 | LPEILGDSQHADVGK | |
| H2AX | P16104 | S139 | GKKATQASQEY____ | ? |
| IKKg (IKBKG, NEMO) | Q9Y6K9 | S85 | ELLHFQASQREEKEF | |
| LKB1 (STK11) | Q15831 | T363 | IEDDIIYTQDFTVPG | + |
| MCM3 | P25205 | S535 | GKKATQASQEY____ | |
| MCM3 | P25205 | S728 | HTPKTADSQETKESQ | |
| MDM2 | Q00987 | S395 | SQESEDYSQPSTSSS | |
| MDM4 | O15151 | S403 | DLAHSSESQETISSM | |
| MRE11A | P49959 | S264 | EQQLFYISQPGSSVV | |
| NBS1 | O60934 | S278 | VDTGITNSQTLIPDC | |
| NBS1 | O60934 | S343 | TTPGPSLSQGVSVDE | |
| NBS1 | O60934 | S397 | EQKFRMLSQDAPTVK | |
| NBS1 | O60934 | S615 | VPESSKISQENEIGK | |
| NFAT5 | O94916 | S1197 | HIQTPMLSQEQAQPP | |
| NFAT5 | O94916 | S1247 | AMQSNSPSQEQQQQQ | |
| NFAT5 | O94916 | S1367 | LVQGSPSSQEQQVTL | |
| p53 | P04637 | S15 | PSVEPPLSQETFSDL | + |
| p53 | P04637 | S20 | PLSQETFSDLWKLLP | + |
| p53 | P04637 | S37 | NVLSPLPSQAMDDLM | + |
| p53 | P04637 | S392 | FKTEGPDSD______ | + |
| p53 | P04637 | S46 | AMDDLMLSPDDIEQW | + |
| p53 | P04637 | S6 | __MEEPQSDPSVEPP | + |
| p53 | P04637 | S9 | EEPQSDPSVEPPLSQ | |
| p63 | Q9H3D4 | S479 | MNKLPSVSQLINPQQ | - |
| PPP1R2 | P41236 | S43 | DEELSKKSQKWDEMN | |
| Rad17 | O75943 | S635 | ETWSLPLSQNSASEL | |
| Rad17 | O75943 | S645 | SASELPASQPQPFSA | |
| Rad9 | Q99638 | S272 | LSDTDSHSQDLGSPE | + |
| RASSF1 | Q9NS23 | S135 | EWETPDLSQAEIEQK | |
| RFWD2 | Q8NHY2 | S387 | SDDSRTASQLDEFQE | |
| RPA2 | P15927 | T21 | YGGAGGYTQSPGGFG | |
| Smc1 | Q14683 | S957 | ISQEEGSSQGEDSVS | |
| Smc1 | Q14683 | S966 | GEDSVSGSQRISSIY | |
| TAO1 (TAOK1) | Q7L7X3 | S990 | SRSTSVTSQISNGSH | + |
| TAO1 (TAOK1) | Q7L7X3 | T643 | EELNKRQTQKDLEHA | + |
| TAO1 (TAOK1) | Q7L7X3 | T785 | SINEMLSTQALRLDE | + |
| TERF1 | P54274 | S219 | SKLLMIISQKDTFHS | ? |
| USP28 | Q96RU2 | S67 | DERVKEPSQDTVATE | |
| USP28 | Q96RU2 | S714 | ESSTNSSSQDYSTSQ | |
| WRN | Q14191 | S1141 | PEKAYSSSQPVISAQ | |
| XPA | P23025 | S173 | VKKNPHHSQWGDMKL | |
| XPA | P23025 | S196 | RSLEVWGSQEALEEA | |
| ZNF148 | Q9UQR1 | S202 | GEKPFQCSQCDMRFI |
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, brain disorders
Specific Diseases (Non-cancerous):
Ataxia Telangiectasia (AT); Ataxia; Nijmegen breakage syndrome; Idiopathic acute transverse myelitis; Cervical dystonia; Telangiectasis; Asphyxiating thoracic dystrophy; Nijmegen breakage syndrome-like disorder; Chromosome 11q Deletion; Ataxia-Telangiectasia variant; Combined Cervical Dystonia;
Comments:
Loss of function mutations in ATM are responsible for ataxia telangiectasia (AT) and immune deficiency. AT is a rare recessive disorder characterized by progressive loss of motor control (from cerebellar ataxia), and dilation of superficial blood vessels in the conjunctiva and eyeballs. About 30% of ATM patients have a strong predisposition to cancer, particularly lymphomas and leukemias. Cells from AT cancer patients are very sensitive to damage by ionizing radiation and following irradiation are resistant to inhibition of DNA synthesis. Many different mutations in AT cancer patients are observed, which is consistent with loss of function mutations that underlie the development of cancer.
Specific Cancer Types:
Breast cancer; mantle cell lymphoma (MCL); chronic lymphocytic leukemia (CLL); B-cell chronic lymphocytic leukemia (B-CLL); bilateral breast cancer; prolymphocytic leukemia (PLL); ovarian cancer; T-cell prolymphocytic leukemia (T-CLL); familial chronic lymphocytic leukemia; breast cancer, somatic; lymphoma, B-cell non-Hodgkin, somatic; T-cell prolymphocytic leukemia, somatic;
Comments:
ATM appears to be a tumour suppressor protein (TSP). Cancer-related mutations in human tumours point to a loss of function of the protein kinase. The active form of the protein kinase normally acts to inhibit tumour cell proliferation. Approximately 30% of cancers, mostly lymphomas and leukemias, arise due to defects in DNA damage repair. ATM mutations are also associated with melanoma, breast, lymphoid tissue, lung, ovarian, stomach, and upper aerodigestive tract tumours. In human tumours, a wide range of point mutations, complex mutations, deletions and insertions are distributed through out the protein, which is characteristic for a tumour suppressor protein.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +48, p<0.051); Brain glioblastomas (%CFC= -59, p<0.043); Brain oligodendrogliomas (%CFC= -72, p<0.023); Cervical cancer stage 2B (%CFC= -59, p<0.051); Classical Hodgkin lymphomas (%CFC= -47, p<0.015); Clear cell renal cell carcinomas (cRCC) (%CFC= +50, p<0.066); Colorectal adenocarcinomas (early onset) (%CFC= +69, p<0.013); and T-cell prolymphocytic leukemia (%CFC= -48, p<0.0001). The COSMIC website notes an up-regulated expression score for ATM in diverse human cancers of 279, which is 0.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 67 for this protein kinase in human cancers was 1.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.14 % in 30040 diverse cancer specimens. This rate is 1.85-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.64 % in 1284 large intestine cancers tested; 0.31 % in 4073 haematopoietic and lymphoid cancers tested; 0.24 % in 859 skin cancers tested; 0.2 % in 619 stomach cancers tested; 0.19 % in 603 endometrium cancers tested; 0.18 % in 500 urinary tract cancers tested; 0.15 % in 2143 lung cancers tested; 0.1 % in 984 upper aerodigestive tract cancers tested; 0.1 % in 1284 liver cancers tested; 0.09 % in 817 prostate cancers tested; 0.09 % in 1333 pancreas cancers tested; 0.08 % in 607 oesophagus cancers tested; 0.07 % in 2139 breast cancers tested; 0.07 % in 1711 kidney cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: N1983S (23); R337C (19); R337H (12); D1853N (13).
Comments:
Broad distribution of mutation sites with point mutations, complex mutations, insertions and deletions over entire protein length. This is characteristic for a tumour suppressor protein.

