Nomenclature
Short Name:
YES
Full Name:
Proto-oncogene tyrosine-protein kinase Yes
Alias:
- c-Yes
- EC 2.7.10.2
- HsT441
- p61-Yes
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Src
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
60801
# Amino Acids:
543
# mRNA Isoforms:
1
mRNA Isoforms:
60,801 Da (543 AA; P07947)
4D Structure:
NA
1D Structure:
Subfamily Alignment
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Palmitoylated:
C3 (predicted).
Serine phosphorylated:
S11, S26, S28, S30, S40, S45, S46, S111, S144, S187, S195, S197, S276, S355.
Myristoylated:
G2.
Threonine phosphorylated:
T21, T37, T38, T190, T225, T226, T427+, T531, T533.
Tyrosine phosphorylated:
Y16, Y31, Y32, Y100, Y141, Y146, Y159, Y194, Y222, Y223, Y336, Y345, Y426+, Y446, Y489, Y537-.
Ubiquitinated:
K191, K235, K437.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 73
73
799
42
868
 7
7
79
23
54
 49
49
538
13
503
 42
42
464
134
699
 80
80
878
39
745
 17
17
183
130
240
 30
30
332
46
426
 96
96
1052
62
1510
 68
68
746
27
650
 20
20
213
114
363
 37
37
406
41
525
 68
68
747
234
699
 17
17
181
46
367
 14
14
157
18
146
 40
40
440
32
529
 13
13
145
22
135
 14
14
153
137
263
 41
41
451
27
651
 8
8
90
131
137
 63
63
685
165
749
 38
38
418
29
646
 44
44
483
35
663
 44
44
482
24
588
 37
37
404
27
484
 21
21
226
29
379
 100
100
1092
87
2432
 15
15
162
52
296
 28
28
301
27
434
 36
36
388
27
516
 4
4
46
42
45
 46
46
504
30
565
 42
42
461
45
388
 32
32
352
86
646
 68
68
743
83
690
 12
12
127
48
176
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 94.2
94.2
94.2
100 99.6
99.6
99.8
100 -
-
-
97 -
-
-
100 97.6
97.6
97.9
98 -
-
-
- 95.9
95.9
97
96 73.6
73.6
83.2
95 -
-
-
- 81.4
81.4
82.6
- 91.9
91.9
96.3
92 89.6
89.6
93.7
91 85.9
85.9
91.3
86 -
-
-
- 54.3
54.3
67.2
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PRODH2 - Q9UF12 |
| 2 | NPHS1 - O60500 |
| 3 | CD36 - P16671 |
| 4 | FLT1 - P17948 |
| 5 | OCLN - Q16625 |
| 6 | ITGB4 - P16144 |
| 7 | PTPRE - P23469 |
| 8 | FAS - P25445 |
| 9 | PDGFRB - P09619 |
| 10 | CDKN1B - P46527 |
| 11 | SLC9A3R1 - O14745 |
| 12 | CD46 - P15529 |
| 13 | MST1R - Q04912 |
| 14 | TRPV4 - Q9HBA0 |
| 15 | GP6 - Q9HCN6 |
Regulation
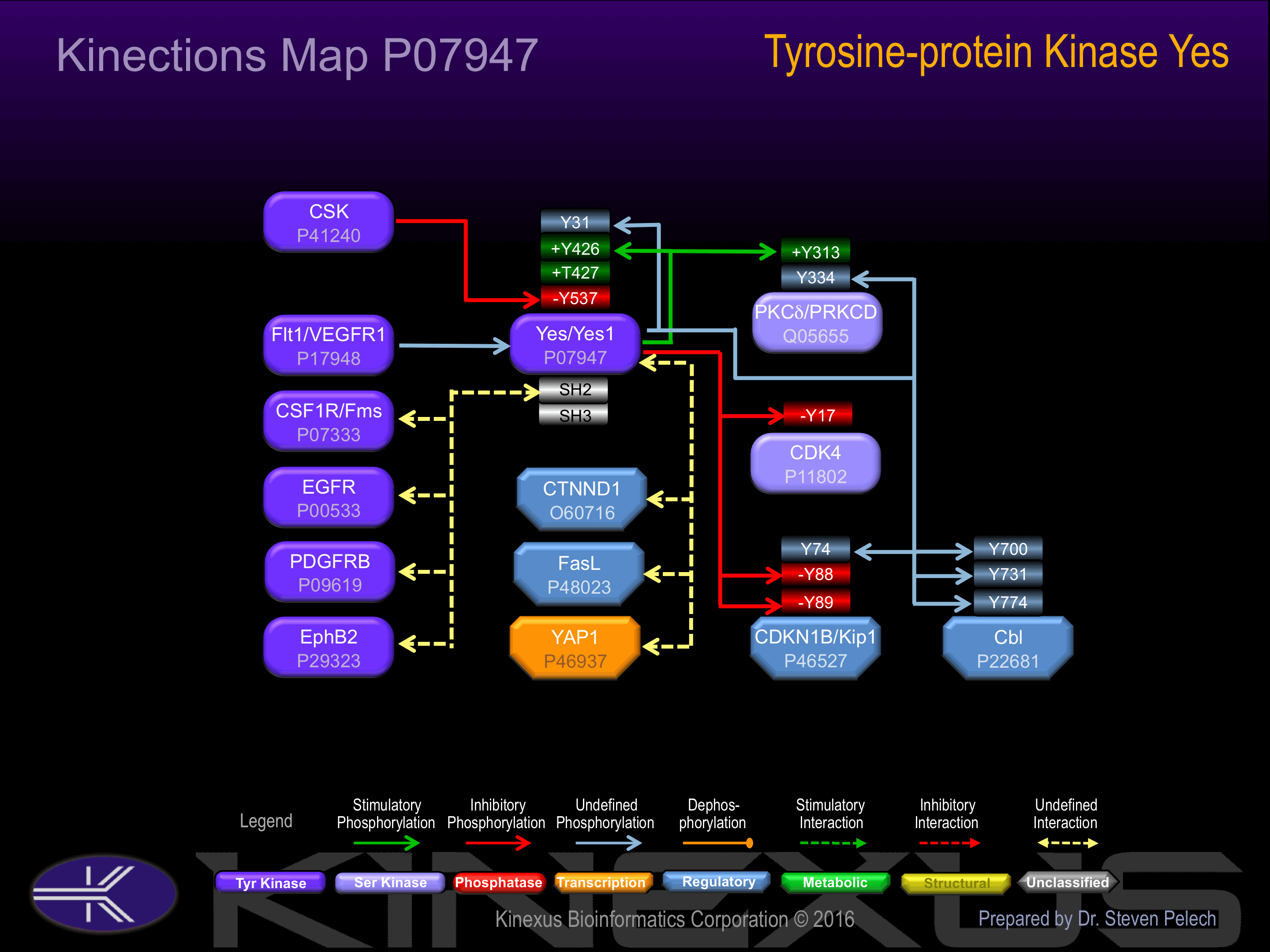
Activation:
Phosphorylation of Tyr-537 increases phosphoransferase activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Cbl | P22681 | Y700 | EGEEDTEYMTPSSRP | |
| Cbl | P22681 | Y731 | QQIDSCTYEAMYNIQ | |
| Cbl | P22681 | Y774 | SENEDDGYDVPKPPV | |
| CDK4 | P11802 | Y17 | AEIGVGAYGTVYKAR | - |
| p27Kip1 | P46527 | Y74 | HKPLEGKYEWQEVEK | ? |
| p27Kip1 | P46527 | Y88 | KGSLPEFYYRPPRPP | - |
| p27Kip1 | P46527 | Y89 | GSLPEFYYRPPRPPK | - |
| PKCd (PRKCD) | Q05655 | Y313 | SSEPVGIYQGFEKKT | + |
| PKCd (PRKCD) | Q05655 | Y334 | MQDNSGTYGKIWEGS | ? |
| Yes | P07947 | Y31 | VSTSVSHYGAEPTTV | |
| Yes | P07947 | Y426 | RLIEDNEYTARQGAK | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
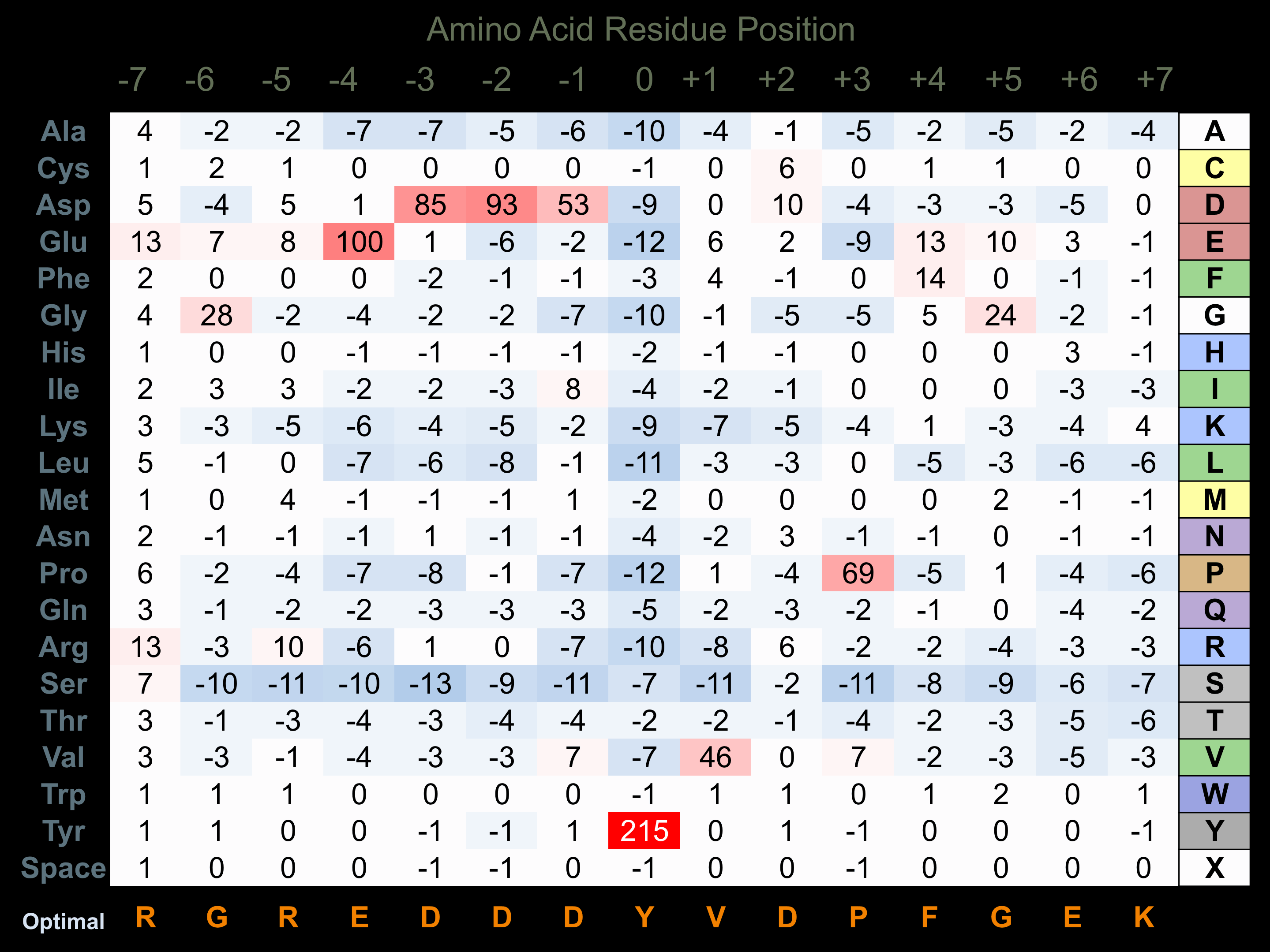
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, gastrointestinal disorders
Specific Diseases (Non-cancerous):
Megaesophagus
Specific Cancer Types:
Follicular lymphomas; Sarcomas
Comments:
Yes may be an oncoprotein (OP) based on its similarity to other Src family protein-tyrosine kinases. Yes is the cellular homologue of the Yamaguchi sarcoma virus oncoprotein. By isotopic in situ hybridization, Yes1 gene localization is frequently associated with chromosome translocation and pathogenesis of follicular lymphoma.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +123, p<(0.0003); Brain glioblastomas (%CFC= -61, p<0.008); Brain oligodendrogliomas (%CFC= -58, p<0.031); Head and neck squamous cell carcinomas (HNSCC) (%CFC= +45, p<0.043); and Oral squamous cell carcinomas (OSCC) (%CFC= +59, p<0.002). The COSMIC website notes an up-regulated expression score for Yes in diverse human cancers of 569, which is 1.2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 116 for this protein kinase in human cancers was 1.9-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. A Y426F mutation in Yes leads to about a 50% reduction in CSK-mediated inhibition of its phosphotransferase activity.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25126 diverse cancer specimens. This rate is only -8 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Frequency of Mutated Sites:
None > 4 in 20,409 cancer specimens
Comments:
Only 2 deletions, and no insertions or complex mutations are noted on the COSMIC website.