Nomenclature
Short Name:
FRAP1
Full Name:
FKBP12-rapamycin complex-associated protein
Alias:
- DJ576K7.1
- FLJ44809
- FRAP
- FRAP2
- RAPT1
- FK506 binding protein 12-rapamycin associated protein 1
- FK506 binding protein 12-rapamycin associated protein 2
- FK506-binding protein 12-rapamycin complex-associated protein 1
- FKBP12-rapamycin complex-associated protein 1
- FKBP-rapamycin associated protein
Classification
Type:
Protein-serine/threonine kinase
Group:
Atypical
Family:
PIKK
SubFamily:
FRAP
Specific Links
Structure
Mol. Mass (Da):
288,892
# Amino Acids:
2549
# mRNA Isoforms:
1
mRNA Isoforms:
288,892 Da (2549 AA; P42345)
4D Structure:
Interacts with PPAPDC3 By similarity. Part of the mammalian target of rapamycin complex 1 (mTORC1) which contains MTOR/FRAP1, MLST8, RPTOR and AKT1S1. mTORC1 binds to and is inhibited by FKBP12-rapamycin. Part of the mammalian target of rapamycin complex
3D Structure:
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
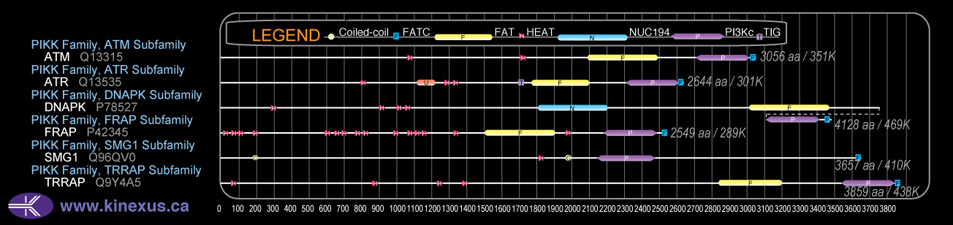
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K1218 (N6), K1256.
Serine phosphorylated:
S92, S567, S719, S909, S918, S1131, S1166, S1171, S1261+, S1288, S1297, S1299, S1415, S1418, S1821, S1859, S1874, S1893, S2155, S2159+, S2442, S2448+, S2450, S2454, S2478, S2481+.
Threonine phosphorylated:
T102, T308, T314, T665, T1162, T1172, T1262, T1502, T1870, T1876, T2164+, T2380, T2434, T2436, T2444, T2446-, T2471, T2473, T2474.
Tyrosine phosphorylated:
Y110, Y1188, Y1232, Y1880, Y2449.
Ubiquitinated:
K243, K251, K898, K900, K1218, K1256, K1293, K1406, K1655, K2066.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-

 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
99 -
-
-
- -
-
-
99.5 -
-
-
99 99.4
99.4
99.5
99 -
-
-
- 98.9
98.9
99.2
99 98.9
98.9
99.2
99 -
-
-
- 88.8
88.8
91.2
- 93.7
93.7
95.4
97 -
-
-
89 89.8
89.8
94.5
91 -
-
-
- 54
54
69.8
56 62
62
75.7
- 34.7
34.7
53.4
51 -
-
-
- -
-
-
- -
-
-
- -
-
-
53 -
-
-
46 40.6
40.6
58.9
46 -
-
-
49
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
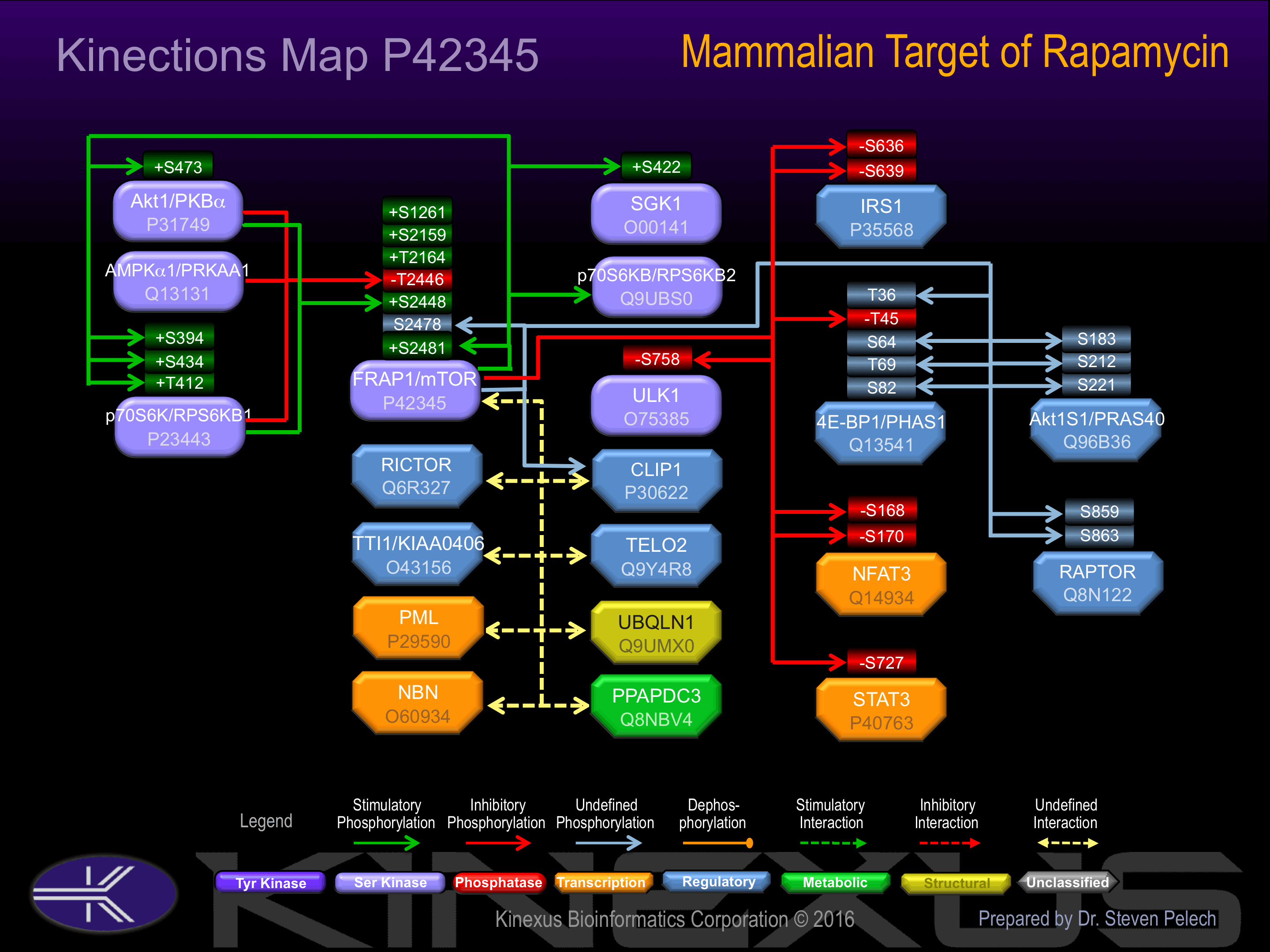
Phosphorylation of Ser-2448 and Ser-2481 increases phosphotransferase activity. Kinase activity is positively regulated by RHEB
Inhibition:
Phosphorylation of Thr-2446 inhibits phosphorylation of the S2448 activatory site. Negatively regulated by DEPDC6.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| 4E-BP1 (EIF4EBP1) | Q13541 | S64 | FLMECRNSPVTKTPP | |
| 4E-BP1 (EIF4EBP1) | Q13541 | S82 | PTIPGVTSPSSDEPP | |
| 4E-BP1 (EIF4EBP1) | Q13541 | T36 | PPGDYSTTPGGTLFS | |
| 4E-BP1 (EIF4EBP1) | Q13541 | T45 | GGTLFSTTPGGTRII | - |
| 4E-BP1 (EIF4EBP1) | Q13541 | T69 | RNSPVTKTPPRDLPT | |
| Akt1 (PKBa) | P31749 | S473 | RPHFPQFSYSASGTA | + |
| Akt1S1 | Q96B36 | S183 | PTQQYAKSLPVSVPV | |
| Akt1S1 | Q96B36 | S212 | EENGPPSSPDLDRIA | |
| Akt1S1 | Q96B36 | S221 | DLDRIAASMRALVLR | |
| FRAP1 (mTOR) | P42345 | S2478 | TGTTVPESIHSFIGD | |
| FRAP1 (mTOR) | P42345 | S2481 | TVPESIHSFIGDGLV | + |
| IRS1 | P35568 | S636 | SGDYMPMSPKSVSAP | - |
| IRS1 | P35568 | S639 | YMPMSPKSVSAPQQI | - |
| NFAT3 | Q14934 | S168 | QGGGAFFSPSPGSSS | - |
| NFAT3 | Q14934 | S170 | GGAFFSPSPGSSSLS | - |
| p70S6K (RPS6KB1) | P23443 | S394 | TRQTPVDSPDDSTLS | + |
| p70S6K (RPS6KB1) | P23443 | S434 | SFEPKIRSPRRFIGS | + |
| p70S6K (RPS6KB1) | P23443 | T412 | NQVFLGFTYVAPSVL | + |
| Raptor | Q8N122 | S859 | DTSSLTQSAPASPTN | |
| Raptor | Q8N122 | S863 | LTQSAPASPTNKGVH | |
| SGK1 | O00141 | S422 | AEAFLGFSYAPPTDS | + |
| STAT3 | P40763 | S727 | NTIDLPMSPRTLDSL | - |
| ULK1 | O75385 | S758 | PVVFTVGSPPSGSTP | - |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
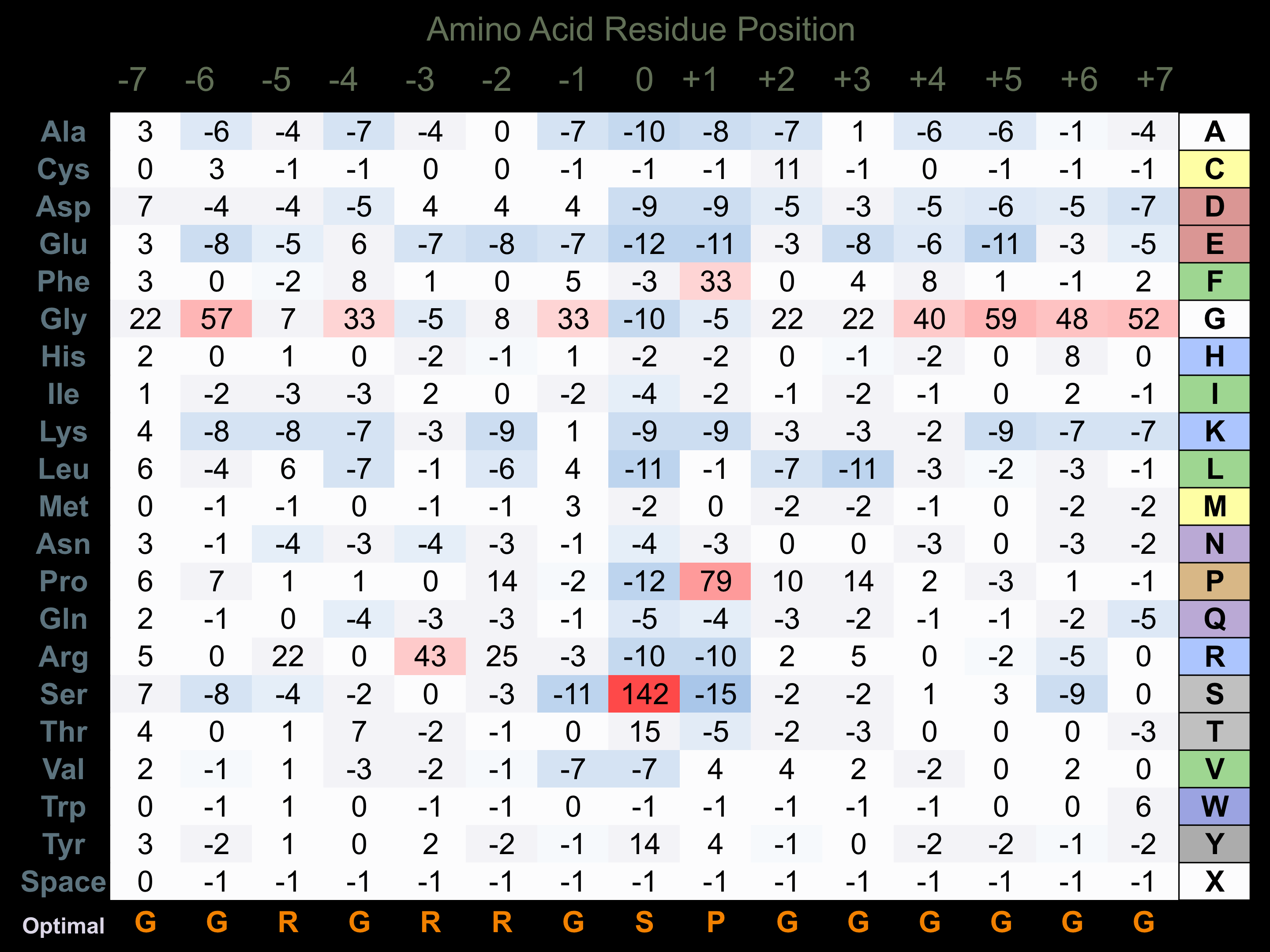
Matrix Type:
Experimentally derived from alignment of 134 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Rapamycin | IC50 100 pM | 5284616 | 413 | 103164548 |
| GSK2126458 | Ki = 180 pM | 25167777 | ||
| Deforolimus | IC50 = 200 pM | 11520894 | 21482695 | |
| Torin1 | IC50 = 290 pM | 49836027 | 1255226 | |
| Everolimus | IC50 = 630 pM | 6442177 | ||
| AZD8055 | IC50 800 pM | 25262965 | 1801204 | 20561789 |
| INK 128 | IC50 = 1 nM | 45375953 | 20622997 | |
| PP242 | Kd = 3 nM | 25243800 | 22037378 | |
| PP121 | IC50 = 10 nM | 24905142 | 18849971 | |
| GSK1059615 | IC50 = 12 nM | 23582824 | 19589091 | |
| PI-103 | Kd = 12 nM | 16739368 | 538346 | 22037378 |
| PF-04691502 | Ki = 16 nM | 25033539 | 21750219 | |
| NVP-BEZ235 | IC50 = 21 nM | 11977753 | 1879463 | 21966435 |
| Wortmannin | IC50 = 40 nM | 312145 | 428496 | 16143526 |
| XL765 | IC50 = 157 nM | 49867926 | ||
| GDC-0941 | Kd = 200 nM | 17755052 | 521851 | 22037378 |
| ZSTK474 | IC50 = 350 nM | 11647372 | 19589091 | |
| TG100115 | Kd = 620 nM | 10427712 | 230011 | 22037378 |
| Ponatinib | IC50 > 1 µM | 24826799 | 20513156 | |
| Silmitasertib | IC50 > 1 µM | 24748573 | 21174434 | |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| Temsirolimus | IC50 = 1.76 µM | 24847874 | 21438579 | |
| LY294002 | IC50 = 2.5 µM | 3973 | 98350 | 15658870 |
Disease Linkage
General Disease Association:
Cancer, eye, neurological, and immune disorders
Specific Diseases (Non-cancerous):
Alexander disease (ALX); Hemimegalencephaly; Paronychia
Comments:
Alexander Disease (ALX) is a rare disease which affects the brain. In ALX the white matter is destroyed and protein deposits (rosenthal fibers) replace the tissue, leading to speech abnormalities, difficulty swallowing, and lack of coordination in sufferers. Hemimegalencephaly is characterized by the enlargement of one side of the brain leading to an oversized and usually malfunctioning brain. Common symptoms in Hemimegalencephaly include impaired development, seizures, brain dysfunction, and spinal cord dysfunction. Paronychia is characterized by bacterial or fungal growth on the hand or foot, at the base of the nails.
Specific Cancer Types:
Tuberous Sclerosis (TSC); Subependymal giant cell astrocytoma (SEGA); Lymphangioleiomyomatosis (LAM); Angiomyolipoma; Breast cancer; Tuberous sclerosis complex; Plasmablastic lymphomas (PBL); Ewing's family of tumours; Kidney angiomyolipoma
Comments:
FRAP1 is a known oncoprotein (OP). Cancer-related mutations in human tumours point to a gain of function of the protein kinase. The active form of the protein kinase normally acts to promote tumour cell proliferation. FRAP1 has been linked with Tuberous Sclerosis, which is a rare disease that leads to a benign tumour growth in the brain and other organs. Tuberous Sclerosis often affects the skin, kidney, and brain. Subependymal Giant Cell Astrocytoma (SEGA) is a rare neuronal astrocytoma with a relation to the FSH signalling, and insulin receptor signalling pathways. Lymphangioleiomyomatosis (LAM) is a rare respiratory growth where LAM cells form cysts over time, and damage whichever tissue they reside in. LAM can also affect kidney, and smooth muscle. Angiomyolipoma is a benign tumour of the kidney, but can also affect the liver and lung. Tuberous Sclerosis Complex is characterized by the growth of benign tumours over the sufferer's body and can affect the skin, brain, and kidneys, among other tissues. Plasmablastic Lymphoma (PBL) is a rare cancer related to plasmacytoma and chronic lymphocytic leukemia. PBL can affect B cells, the spinal cord, and the liver. The rare cancer Ewing's Family of tumours is related to medullablastoma and cerebral primitive neuroectodermal tumour. Kidney Angiomyolipoma is related to the PI3K/Akt signalling pathway and the glioblastoma multiform pathway. In FRAP1 the S2159A and T2164A mutations together will lower autophosphorylation of the mTORC1-associated S-2481 residue. RPS6KB1 is phosphorylated more with the S2159D and T2164E mutations.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for FRAP1 in diverse human cancers of 395, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 250 for this protein kinase in human cancers was 4.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. mTOR phosphotransferase activity can be increased with a T2173A mutation. Kinase phosphotransferase activity can be almost completely abrogated with a H2340A mutation. In FRAP1 the S2159A and T2164A mutations together will lower autophosphorylation of the mTORC1-associated S2481 residue. The substrate RPS6KB1 is phosphorylated more with the S2159D and T2164E mutations. The mTOR kinase phosphotransferase activity can be increased with a T2173A mutation. Its kinase phosphotransferase activity can be almost completely removed with a H2340A mutation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25904 diverse cancer specimens. This rate is very similar (+ 8% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.32 % in 1219 large intestine cancers tested; 0.26 % in 807 skin cancers tested; 0.24 % in 602 endometrium cancers tested; 0.21 % in 590 stomach cancers tested; 0.19 % in 1398 kidney cancers tested; 0.13 % in 1757 lung cancers tested; 0.11 % in 500 urinary tract cancers tested; 0.07 % in 605 oesophagus cancers tested; 0.07 % in 1498 breast cancers tested; 0.06 % in 984 upper aerodigestive tract cancers tested; 0.05 % in 905 ovary cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: S2215Y (10);S2215F (5); L2431P (7); R2505P (4); R2505Q (2); R2505L (1); I2500M (5); L2427Q (4); L2427R (3).
Comments:
Nine deletions, and no insertions or complex mutations are noted on the COSMIC website.