Nomenclature
Short Name:
SIK
Full Name:
Serine-threonine-protein kinase SNF1-like kinase 1
Alias:
- EC 2.7.11.1
- Msk
- Serine/threonine-protein kinase SNF1LK
- SN1L1
- SNF1-like kinase
- SNF1LK
- Myocardial SNF1-like kinase
- Salt-inducible kinase
- Salt-inducible kinase 1
- Serine/threonine-protein kinase SNF1-like kinase 1
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
CAMKL
SubFamily:
QIK
Specific Links
Structure
Mol. Mass (Da):
84,902
# Amino Acids:
783
# mRNA Isoforms:
1
mRNA Isoforms:
84,902 Da (783 AA; P57059)
4D Structure:
Binds to and is activated by YWHAZ when phosphorylated on Thr-182
1D Structure:
Subfamily Alignment
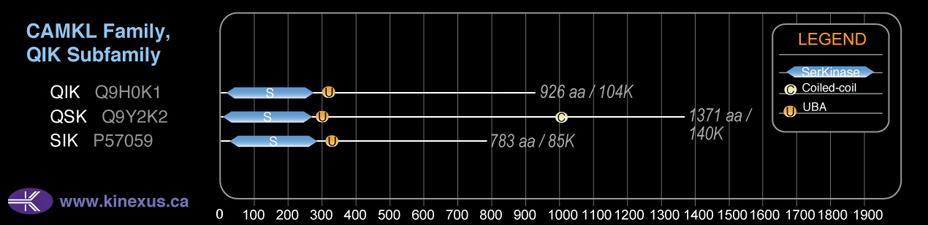
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S186+, S370, S435, S437, S516, S534, S575, S577, S626, S634, S644.
Threonine phosphorylated:
T182+, T460, T473, T479, T577.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 10
10
1025
25
1145
 3
3
337
11
313
 2
2
208
1
0
 6
6
636
69
1627
 8
8
872
24
682
 3
3
265
46
271
 2
2
226
29
483
 5
5
553
20
678
 3
3
327
10
282
 2
2
168
69
221
 3
3
357
14
434
 10
10
1077
100
676
 2
2
158
12
110
 6
6
578
9
547
 1.3
1.3
138
11
170
 10
10
1026
13
1416
 5
5
485
103
359
 2
2
173
8
133
 0.9
0.9
92
61
61
 9
9
928
84
805
 2
2
212
10
161
 2
2
176
12
179
 3
3
291
10
304
 1.2
1.2
119
8
93
 2
2
182
10
184
 13
13
1325
44
1932
 1.1
1.1
111
15
47
 4
4
401
8
254
 2
2
194
8
171
 2
2
223
28
138
 6
6
600
18
407
 100
100
10273
32
13558
 0.4
0.4
38
29
57
 8
8
804
57
779
 4
4
406
35
791
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.6
99.6
99.6
70 -
-
-
- -
-
-
73 -
-
-
- 69.3
69.3
73.6
78 -
-
-
- 81.3
81.3
86.2
80 81.6
81.6
86.3
81 -
-
-
- 70.4
70.4
77
- 66.7
66.7
76.7
66 -
-
-
- 32
32
44.1
- -
-
-
- 28.9
28.9
38
- 36.5
36.5
51
- 24.5
24.5
39.4
- -
-
-
- -
-
-
- -
-
-
- -
-
-
40 28.2
28.2
41
- 27.2
27.2
42.8
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | VAPA - Q9P0L0 |
| 2 | CRTC2 - Q53ET0 |
| 3 | NUAK1 - O60285 |
| 4 | YWHAE - P62258 |
| 5 | YWHAZ - P63104 |
Regulation
Activation:
Phosphorylation at Ser-186 increases phosphotransferase activity and induces interaction with 14-3-3. Also activated by phosphorylation on Thr-182 by STK11 in complex with STE20-related adapter-alpha (STRAD alpha) pseudo kinase and CAB39.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
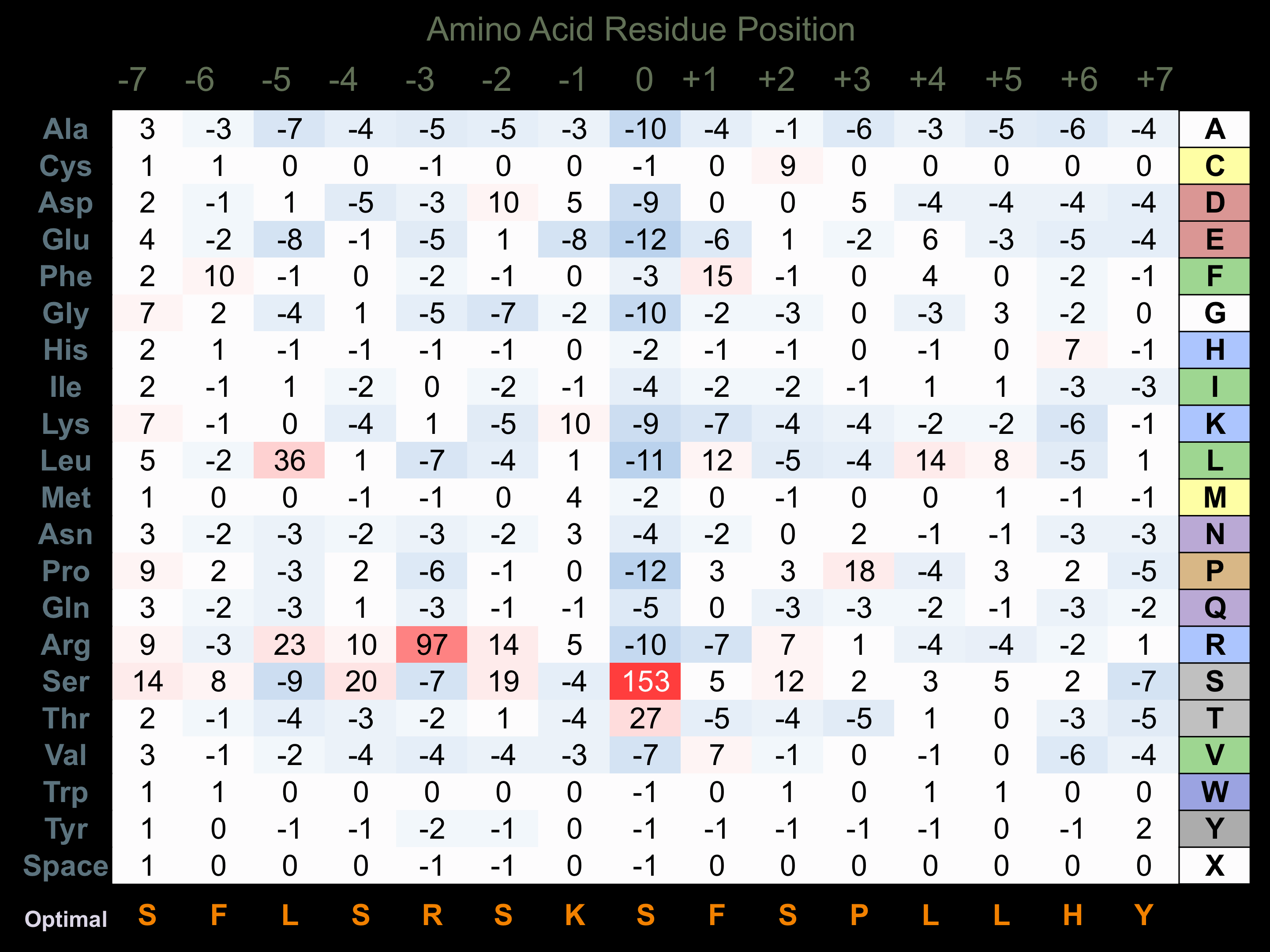
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Breast cancer
Comments:
SIK may be a tumour requiring protein (TRP). The loss of SIK1 has been correlated with poor prognosis for individuals suffering from breast cancer. A K56M mutation can result in loss of kinase phosphotransferase activity. Kinase phosphotransferase activity can be decreased without affecting autophosphorylation levels of SIK through either a S135A, or a S209A, or a S248A mutation. LKB1/LKB1-mediated phosphorylation and activation of SIK can be inhibited through a T182A mutation in SIK. Partial inhibition of phosphorylation and of kinase phosphotransferase activity has been observed with the S186A/D/C/G mutation. Abrogation of autophosphorylation and kinase phosphotransferase activity can occur with a S186T mutation.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= -47, p<0.076); Brain oligodendrogliomas (%CFC= -46, p<0.081); Breast epithelial cell carcinomas (%CFC= -72, p<0.005); Cervical epithelial cancer (%CFC= +85, p<0.032); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= -81, p<0.002); Colorectal adenocarcinomas (early onset) (%CFC= +312, p<0.017); Lung adenocarcinomas (%CFC= -46, p<0.0005); Skin fibrosarcomas (%CFC= -89);Skin melanomas - malignant (%CFC= -79, p<0.007); Uterine fibroids (%CFC= -65, p<0.002); and Uterine leiomyomas from fibroids (%CFC= -75, p<0.06). The COSMIC website notes an up-regulated expression score for SIK in diverse human cancers of 359, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 24737 diverse cancer specimens. This rate is only 12 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.43 % in 864 skin cancers tested; 0.31 % in 1270 large intestine cancers tested; 0.28 % in 942 upper aerodigestive tract cancers tested; 0.16 % in 548 urinary tract cancers tested; 0.13 % in 589 stomach cancers tested; 0.13 % in 1634 lung cancers tested; 0.11 % in 833 ovary cancers tested; 0.11 % in 710 oesophagus cancers tested; 0.09 % in 558 thyroid cancers tested; 0.08 % in 603 endometrium cancers tested; 0.08 % in 1512 liver cancers tested; 0.05 % in 273 cervix cancers tested; 0.05 % in 238 bone cancers tested; 0.05 % in 1276 kidney cancers tested; 0.04 % in 1316 breast cancers tested; 0.03 % in 2082 central nervous system cancers tested; 0.01 % in 881 prostate cancers tested; 0.01 % in 2009 haematopoietic and lymphoid cancers tested; 0.01 % in 1459 pancreas cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: T479P (14); T473P (12); S478P (10).
Comments:
Only 7 deletions, no insertions or complex mutations are noted on the COSMIC website.

