Nomenclature
Short Name:
GSK3B
Full Name:
Glycogen synthase kinase-3 beta
Alias:
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
GSK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
46,744
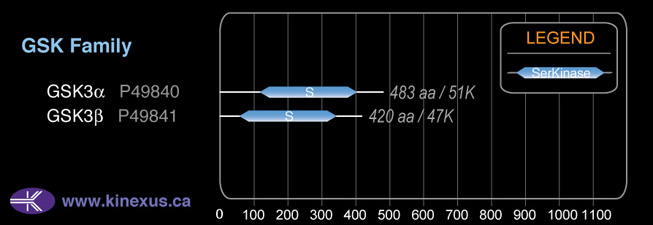
# Amino Acids:
420
# mRNA Isoforms:
2
mRNA Isoforms:
48,034 Da (433 AA; P49841-2); 46,744 Da (420 AA; P49841)
4D Structure:
Monomer By similarity. Interacts with ARRB2 and DISC1 By similarity. Interacts with CABYR, MMP2, MUC1, NIN and PRUNE Interacts with AXIN1; the interaction mediates hyperphosphorylates of CTNNB1 leading to its ubiquitination and destruction. Interacts with and phosphorylates SNAI1.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S9-, S13, S21, S25, S35, S203, S215+, S219-, S389, S417.
Sumoylated:
K292.
Threonine phosphorylated:
T7, T8, T43, T275, T277, T390, T392, T395, T402, T420.
Tyrosine phosphorylated:
Y71, Y114, Y117, Y216+, Y221-, Y222-, Y288.
Ubiquitinated:
K205, K292, K297.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 95
95
1332
37
1161
 9
9
130
15
92
 18
18
255
10
174
 29
29
403
137
428
 70
70
976
41
635
 20
20
277
72
442
 21
21
294
53
493
 100
100
1404
41
3494
 49
49
685
10
639
 12
12
165
142
135
 12
12
173
29
210
 41
41
576
168
624
 10
10
142
21
139
 5
5
64
12
41
 9
9
124
25
132
 7
7
104
24
72
 17
17
236
274
1388
 17
17
236
19
197
 15
15
205
112
192
 62
62
869
140
620
 10
10
134
25
142
 11
11
156
27
156
 14
14
196
19
169
 29
29
414
19
421
 13
13
176
25
166
 87
87
1222
93
2529
 13
13
177
24
201
 12
12
171
19
185
 13
13
178
19
186
 9
9
130
56
110
 68
68
956
18
658
 56
56
789
36
1690
 8
8
109
89
143
 65
65
912
109
731
 16
16
231
61
234
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 98.1
98.1
98.8
98 94.8
94.8
96.4
- -
-
-
99 -
-
-
97 99.8
99.8
99.8
100 -
-
-
- 99
99
99
99 98.8
98.8
99
99 -
-
-
- -
-
-
- 93.8
93.8
95.2
97 93.3
93.3
97.1
91 95
95
97.2
96 -
-
-
- 31.9
31.9
35
70 71
71
80.4
- 64.8
64.8
74.3
78 -
-
-
- -
-
-
- -
-
-
- -
-
-
68 60.2
60.2
74
68 44.8
44.8
63.1
54 -
-
-
62
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Phosphorylation at Thr-216 increases phosphotransferase activity and promotes nuclear localization.
Inhibition:
Phosphorylation at Ser-9 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| GSK3B | P49841 | S9 | SGRPRTTSFAESCKP | - |
| PKCh | P24723 | S9 | SGRPRTTSFAESCKP | - |
| PKCg | P05129 | S9 | SGRPRTTSFAESCKP | - |
| PKCb | P05771 | S9 | SGRPRTTSFAESCKP | - |
| PKCd | Q05655 | S9 | SGRPRTTSFAESCKP | - |
| PKCb | P05771 | S9 | SGRPRTTSFAESCKP | - |
| RSK2 | P51812 | S9 | SGRPRTTSFAESCKP | - |
| RSK1 | Q15418 | S9 | SGRPRTTSFAESCKP | - |
| KHS1 | Q9Y4K4 | S9 | SGRPRTTSFAESCKP | - |
| PKCz | Q05513 | S9 | SGRPRTTSFAESCKP | - |
| ILK | Q13418 | S9 | SGRPRTTSFAESCKP | - |
| PKG1 | Q13976 | S9 | SGRPRTTSFAESCKP | - |
| PKCa | P17252 | S9 | SGRPRTTSFAESCKP | - |
| p70S6K | P23443 | S9 | SGRPRTTSFAESCKP | - |
| Akt1 | P31749 | S9 | SGRPRTTSFAESCKP | - |
| AKT3 | Q9Y243 | S9 | SGRPRTTSFAESCKP | - |
| PKACa | P17612 | S9 | SGRPRTTSFAESCKP | - |
| AurA | O14965 | S9 | SGRPRTTSFAESCKP | - |
| ERK2 | P28482 | T43 | KVTTVVATPGQGPDR | |
| ERK1 | P27361 | T43 | KVTTVVATPGQGPDR | |
| GSK3B | P49841 | Y216 | RGEPNVSYICSRYYR | + |
| MEK1 | Q02750 | Y216 | RGEPNVSYICSRYYR | + |
| MEK2 | P36507 | Y216 | RGEPNVSYICSRYYR | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| PPP1R16B | Q96T49 | S333 | LRHKSSLSRRTSSAG | |
| PPP1R16B | Q96T49 | S337 | SSLSRRTSSAGSRGK | |
| PPP1R2 | P41236 | T72 | MKIDEPSTPYHSMMG | |
| PSEN1 | P49768 | S353 | SHLGPHRSTPESRAA | |
| PSEN1 | P49768 | S357 | PHRSTPESRAAVQEL | |
| PTEN | P60484 | S362 | SNPEASSSTSVTPDV | |
| PTEN | P60484 | T366 | ASSSTSVTPDVSDNE | - |
| PXN | P49023 | S126 | SFPNKQKSAEPSPTV | |
| SFPQ (PSF) | P23246 | T687 | PRGMGPGTPAGYGRG | |
| SIK | P57059 | S186 | PLSTWCGSPPYAAPE | + |
| SIK | P57059 | T182 | KSGEPLSTWCGSPPY | + |
| SNAI1 | O95863 | S100 | DEDSGKGSQPPSPPS | |
| SNAI1 | O95863 | S104 | GKGSQPPSPPSPAPS | |
| SNAI1 | O95863 | S107 | SQPPSPPSPAPSSFS | |
| SNAI1 | O95863 | S111 | SPPSPAPSSFSSTSV | |
| SNAI1 | O95863 | S115 | PAPSSFSSTSVSSLE | |
| SNAI1 | O95863 | S119 | SFSSTSVSSLEAEAY | |
| SNAI1 | O95863 | S96 | TSLSDEDSGKGSQPP | |
| SNCAIP | Q9Y6H5 | S556 | AQKSEGKSLPSSPSS | |
| SPAG5 | Q96R06 | S974 | EESLAEMSIMTTELQ | |
| SPAG5 | Q96R06 | T111 | PIPQISSTPKTSEEA | |
| SPAG5 | Q96R06 | T937 | ADEEPESTPVPLLGS | |
| SPAG5 | Q96R06 | T978 | AEMSIMTTELQSLCS | |
| SREBP1 | P36956 | S430 | DTLTPPPSDAGSPFQ | |
| SREBP1 | P36956 | S434 | PPPSDAGSPFQSSPL | |
| SREBP1 | P36956 | T426 | TEVEDTLTPPPSDAG | |
| Tau | P10636 | S515 | GDRSGYSSPGSPGTP | |
| Tau | P10636 | S518 | SGYSSPGSPGTPGSR | |
| Tau | P10636 | S551 | VVRTPPKSPSSAKSR | |
| Tau | P10636 | S578 | NVKSKIGSTENLKHQ | |
| Tau | P10636 | S720 | PVVSGDTSPRHLSNV | |
| Tau | P10636 | S729 | RHLSNVSSTGSIDMV | |
| Tau | P10636 | S738 | GSIDMVDSPQLATLA | |
| Tau | P10636 | T497 | KTPPAPKTPPSSGEP | |
| Tau | P10636 | T521 | SSPGSPGTPGSRSRT | |
| Tau | P10636 | T528 | TPGSRSRTPSLPTPP | |
| Tau | P10636 | T533 | SRTPSLPTPPTREPK | |
| Tau | P10636 | T547 | KKVAVVRTPPKSPSS | |
| Tau iso8 | P10636-8 | S214 | GSRSRTPSLPTPPTR | |
| Tau iso8 | P10636-8 | S396 | GAEIVYKSPVVSGDT | |
| Tau iso8 | P10636-8 | S400 | VYKSPVVSGDTSPRH | |
| Tau iso8 | P10636-8 | S409 | DTSPRHLSNVSSTGS | |
| Tau iso9 (Tau-F) | P10636-9 | S356 | RVQSKIGSLDNITHV | |
| TNKS | O95271 | S978 | VVSASLISPASTPSC | |
| TNKS | O95271 | S987 | ASTPSCLSAASSIDN | |
| TNKS | O95271 | S991 | SCLSAASSIDNLTGP | |
| TNKS | O95271 | T982 | SLISPASTPSCLSAA | |
| TSC2 | P49815 | S1375 | GRPSVDLSFQPSQPL | |
| TSC2 | P49815 | S1379 | VDLSFQPSQPLSKSS | |
| TSC2 | P49815 | S1383 | FQPSQPLSKSSSSPE | |
| TSC2 | P49815 | T1373 | SSEGARPTVDLSFQP | |
| VHL | P40337 | S68 | PVLRSVNSREPSQVI | |
| APP | P05067 | T743 | VEVDAAVTPEERHLS | |
| Ataxin-3 | P54252 | S256 | LRRAIQLSMQGSSRN | |
| Axin | O15169 | S486 | LRTPGRQSPGPGHRS | |
| Axin | O15169 | T481 | HVQRVLRTPGRQSPG | |
| Baxa (Bax) | Q07812 | S163 | GGWDGLLSYFGTPTW | |
| BCAM (Lu) | P50895 | S596 | PPGEPGLSHSGSEQP | |
| BCLAF1 | Q9NYF8 | S531 | KSTFREESPLRIKMI | |
| C-EBPa | P49715 | T226 | HLQPGHPTPPPTPVP | |
| C-EBPb | P17676 | T235 | SSSSPPGTPSPADAK | |
| C/EBPb | P17676 | S231 | LSTSSSSSPPGTPSP | |
| C/EBPb | P17676 | T226 | GSSGSLSTSSSSSPP | |
| Cdc25A | P30304 | S76 | SNLQRMGSSESTDSG | |
| CdGAP | Q9ULL6 | T790 | PPAPPPPTPLEESTP | |
| CDH1 | P12830 | S847 | SEAASLSSLNSSESD | |
| CDX2 | Q99626 | S283 | RSVPEPLSPVSSLQA | |
| CIITA | P33076 | S373 | VQARLERSSSKSLER | |
| Cip1 (p21, CDKN1A) | P38936 | S113 | EEDHVDLSLSCTLVP | |
| Cip1 (p21, CDKN1A) | P38936 | T56 | NFDFVTETPLEGDFA | |
| CREB1 | P16220 | S129 | QKRREILSRRPSYRK | + |
| CRMP2 (DPYSL2) | Q16555 | S518 | KTVTPASSAKTSPAK | |
| CRMP2 (DPYSL2) | Q16555 | T509 | PVCEVSVTPKTVTPA | |
| CRMP2 (DPYSL2) | Q16555 | T514 | SVTPKTVTPASSAKT | |
| CRMP4 | Q6DEN2 | S632 | KGGTPAGSARGSPTR | |
| CRMP4 | Q6DEN2 | T623 | PVFDLTTTPKGGTPA | |
| CRMP4 | Q6DEN2 | T628 | TTTPKGGTPAGSARG | |
| CRY2 | Q8IV71 | S554 | AGPRPLPSGPASPKR | |
| CRY2 | Q8IV71 | S558 | PLPSGPASPKRKLEA | |
| CTNNB1 | P35222 | S33 | QQQSYLDSGIHSGAT | - |
| CTNNB1 | P35222 | S37 | YLDSGIHSGATTTAP | ? |
| CTNNB1 | P35222 | S45 | GATTTAPSLSGKGNP | + |
| CTNNB1 | P35222 | T41 | GIHSGATTTAPSLSG | ? |
| CTNND1 | O60716 | S252 | MEGYRAPSRQDVYGP | |
| CTNND1 | O60716 | T310 | GTARRTGTPSDPRRR | |
| Cyclin D1 (CCND1) | P24385 | T286 | EEVDLACTPTDVRDV | |
| Cyclin D1 (CCND1) | P24385 | T288 | VDLACTPTDVRDVDI | |
| Cyclin D2 (CCND2) | P30279 | T280 | DELDQASTPTDVRDI | |
| Cyclin D3 (CCND3) | P30281 | T283 | QGPSQTSTPTDVTAI | |
| Cyclin E1 (CCNE1) | P24864 | S73 | AVCADPCSLIPTPDK | |
| Cyclin E1 (CCNE1) | P24864 | T395 | PLPSGLLTPPQSGKK | |
| Cyclin E1 (CCNE1) | P24864 | T77 | DPCSLIPTPDKEDDD | |
| DSCR1 | P53805 | S108 | PDKQFLISPPASPPV | |
| DSCR1 | P53805 | S112 | FLISPPASPPVGWKQ | |
| E2F1 | Q01094 | S403 | PEEFISLSPPHEALD | |
| E2F1 | Q01094 | T433 | DCDFGDLTPLDF___ | |
| eIF2B-e | Q13144 | S540 | MDSEEPDSRGGSPQM | - |
| ERa (ESR1) | P03372 | S102 | GGFPPLNSVSPSPLM | + |
| ERa (ESR1) | P03372 | S104 | FPPLNSVSPSPLMLL | + |
| ERa (ESR1) | P03372 | S106 | PLNSVSPSPLMLLHP | + |
| ERa (ESR1) | P03372 | S118 | LHPPPQLSPFLQPHG | + |
| FAK (PTK2) | Q05397 | S722 | PSRPGYPSPRSSEGF | - |
| FBX4 | Q9UKT5 | S12 | EPRSGTNSPPPPFSD | |
| FRAT1 | Q92837 | S188 | RLQQRRGSQPETRTG | |
| GR | P04150 | S404 | SMRPDVSSPPSSSST | - |
| GSK3b | P49841 | S9 | SGRPRTTSFAESCKP | - |
| GSK3b | P49841 | Y216 | RGEPNVSYICSRYYR | + |
| GYS1 | P13807 | S641 | YRYPRPASVPPSPSL | - |
| GYS1 | P13807 | S645 | RPASVPPSPSLSRHS | - |
| GYS1 | P13807 | S649 | VPPSPSLSRHSSPHQ | |
| GYS1 | P13807 | S653 | PSLSRHSSPHQSEDE | |
| GYS2 | P54840 | S641 | FKYPRPSSVPPSPSG | - |
| GYS2 | P54840 | S645 | RPSSVPPSPSGSQAS | - |
| H1B | P16401 | T11 | TAPAETATPAPVEKS | |
| hnRNP D | Q14103 | S83 | DEGHSNSSPRHSEAA | |
| HSF1 | Q00613 | S303 | RVKEEPPSPPQSPRV | |
| HSL | Q05469 | S855 | EPMRRSVSEAALAQP | |
| IGF1R | P08069 | S1278 | EPGFREVSFYYSEEN | - |
| IRS1 | P35568 | S337 | SDGEGTMSRPASVDG | |
| Jun (c-Jun) | P05412 | S243 | PGETPPLSPIDMESQ | - |
| Jun (c-Jun) | P05412 | T239 | VPEMPGETPPLSPID | - |
| L-Myc | E9PQS5 | S38 | KKFELVPSPPTSPPW | |
| L-Myc | E9PQS5 | S42 | LVPSPPTSPPWGLGP | |
| LRP6 | O75581 | S1490 | AILNPPPSPATERSH | |
| MAP1B | P46821 | S1265 | SLSPSPPSPLEKTPL | |
| MAP1B | P46821 | T1270 | PPSPLEKTPLGERSV | |
| MAP2 | P11137 | T1616 | YSSRTPGTPGTPSYP | |
| MAP2 | P11137 | T1619 | RTPGTPGTPSYPRTP | |
| MARK1 | Q9P0L2 | S219 | KLDTFCGSPPYAAPE | - |
| MARK2 | Q7KZI7 | S212 | KLDTFCGSPPYAAPE | - |
| MCL1 | Q07820 | S159 | NNTSTDGSLPSTPPP | |
| MDM2 | Q00987 | S242 | WLDQDSVSDQFSVEF | + |
| MDM2 | Q00987 | S256 | FEVESLDSEDYSLSE | |
| MITF | O75030 | S405 | QARAHGLSLIPSTGL | + |
| MKI67IP (NIFK) | Q9BYG3 | S230 | TPEKTVDSQGPTPVC | |
| MKI67IP (NIFK) | Q9BYG3 | T234 | TVDSQGPTPVCTPTF | ? |
| MUC1 | P15941 | S1227 | PPSSTDRSPYEKVSA | - |
| Myc | P01106 | T58 | KKFELLPTPPLSPSR | |
| MYLK1 (smMLCK) | Q15746 | S1776 | GRKSSTGSPTSPLNA | |
| Myocardin | Q8IZQ8 | S451 | NGFYHFGSTSSSPPI | |
| Myocardin | Q8IZQ8 | S455 | HFGSTSSSPPISPAS | |
| Myocardin | Q8IZQ8 | S459 | TSSSPPISPASSDLS | |
| Myocardin | Q8IZQ8 | S463 | PPISPASSDLSVAGS | |
| Myocardin | Q8IZQ8 | S626 | DQTNVLSSTFLSPQC | |
| Myocardin | Q8IZQ8 | S630 | VLSSTFLSPQCSPQH | |
| Myocardin | Q8IZQ8 | S634 | TFLSPQCSPQHSPLG | |
| Myocardin | Q8IZQ8 | S638 | PQCSPQHSPLGAVKS | |
| NACA | Q13765 | T159 | NIQENTQTPTVQEES | |
| NDRG1 | Q92597 | S342 | TSLDGTRSRSHTSEG | |
| NDRG1 | Q92597 | S352 | HTSEGTRSRSHTSEG | |
| NDRG2 | Q9UN36 | S328 | CMTRLSRSRTASLTS | |
| Neurogenin 2 | Q9H2A3 | S234 | SCTLSPASPGSDVDY | |
| Neurogenin 2 | Q9H2A3 | S239 | SPYSCTLSPASPAGS | |
| NFH | P12036 | S503 | GGEEETKSPPAEEAA | |
| NFkB-p100 | Q00653 | S222 | QPIHDSKSPGASNLK | ? |
| NFkB-p100 | Q00653 | S707 | EPLCPLPSPPTSDSD | ? |
| NFkB-p100 | Q00653 | S711 | PLPSPPTSDSDSDSE | ? |
| NFkB-p105 | P19838 | S903 | KTTSQAHSLPLSPAS | + |
| NFkB-p105 | P19838 | S907 | QAHSLPLSPASTRQQ | + |
| NFkB-p65 | Q04206 | S468 | AVFTDLASVDNSEFQ | |
| NFM (Neurofilament M) | P07197 | S510 | EEVAAKKSPVKATAP | |
| NFM (Neurofilament M) | P07197 | S666 | PVEEKGKSPVSKSPV | |
| NR1D1 | P20393 | S55 | CPTYFPPSPTGSLTQ | + |
| NR1D1 | P20393 | S59 | FPPSPTGSLTQDPAR | + |
| p53 | P04637 | S33 | LPENNVLSPLPSQAM | + |
| p53 | P04637 | S376 | LKSKKGQSTSRHKKL | + |
| PDX1 | P52945 | S61 | LGALEQGSPPDISPY | + |
| PDX1 | P52945 | S66 | QGSPPDISPYEVPPL | + |
| PKCe (PRKCE) | Q02156 | S346 | SEEDRSKSAPTSPCD | ? |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
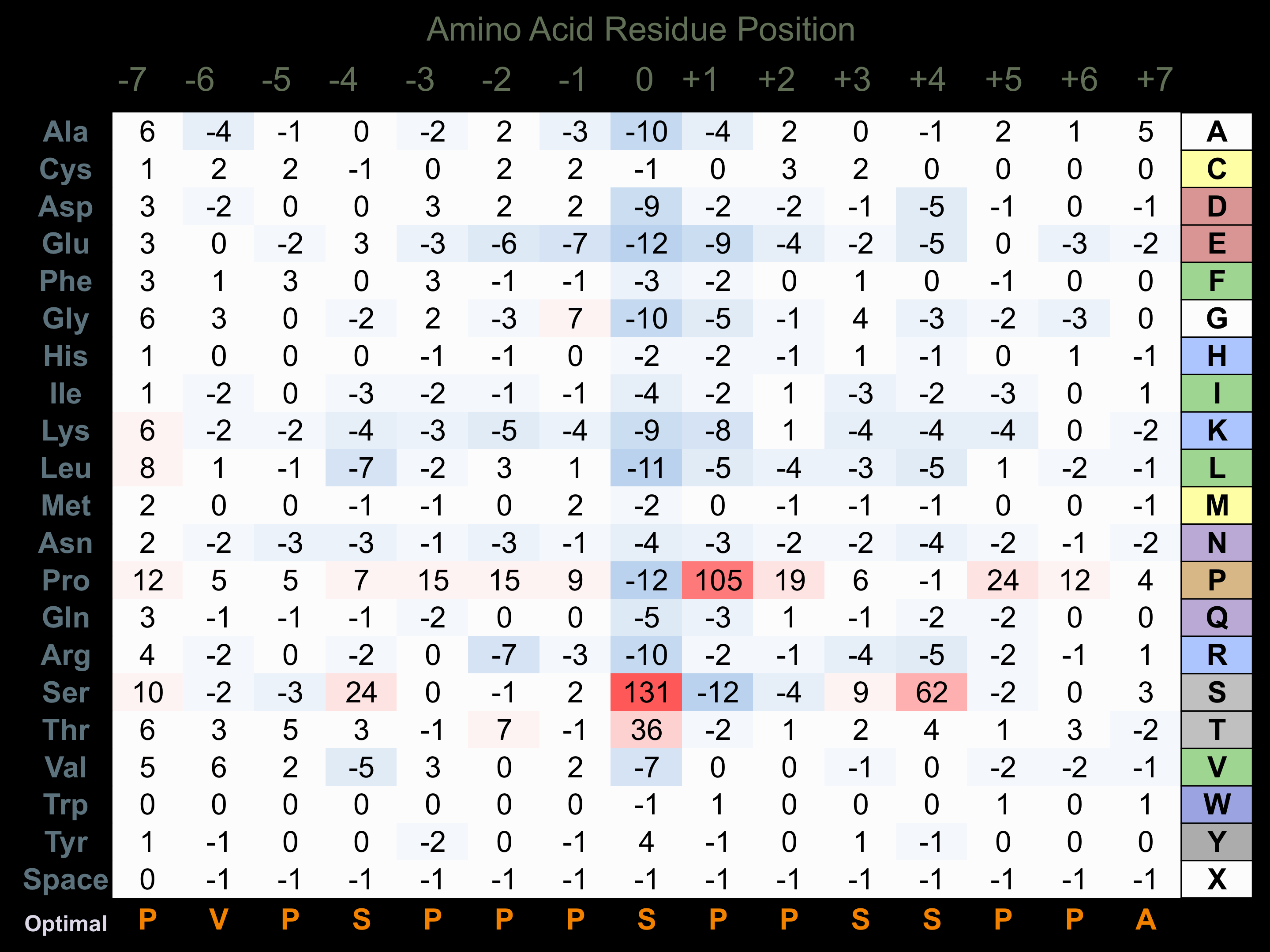
Matrix Type:
Experimentally derived from alignment of 243 known protein substrate phosphosites and 28 peptides phosphorylated by recombinant GSK3-beta in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Neurological disorders
Specific Diseases (Non-cancerous):
Alzheimer's disease (AD); Parkinson's disease; Leopard syndrome; Aneurysmal bone cysts; Waardenburg syndrome Type 2
Comments:
Mutations on GSK3B have been shown to associated with Parkinson disease, and homozygous GSK3B TT haplotype is associated with Alzheimer disease.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +112, p<0.0001); Bladder carcinomas (%CFC= +112, p<0.0001); Bladder carcinomas (%CFC= +52, p<0.085); Brain glioblastomas (%CFC= +308, p<0.005); Cervical cancer (%CFC= -57, p<0.0002); Cervical cancer stage 1B (%CFC= +56, p<0.073); Cervical cancer stage 2B (%CFC= +73, p<0.05); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +104, p<0.058); Prostate cancer - metastatic (%CFC= +95, p<0.0001); Prostate cancer - primary (%CFC= -60, p<0.0001); Prostate cancer - primary (%CFC= +63, p<0.011); Skin melanomas - malignant (%CFC= +59, p<0.0002); and Uterine leiomyomas (%CFC= -54, p<0.016).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 24865 diverse cancer specimens. This rate is only 16 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Frequency of Mutated Sites:
None > 4 in 20,655 cancer specimens
Comments:
No deletions, insertions or complex mutations are noted on the COSMIC website.

