Nomenclature
Short Name:
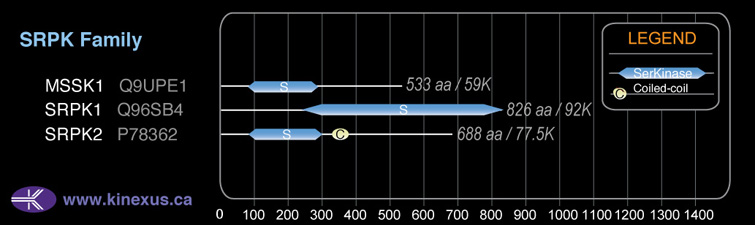
SRPK2
Full Name:
Serine kinase SRPK2
Alias:
- EC 2.7.11.1
- FLJ36101
- SR-protein-specific kinase 2
- Serine/arginine-rich protein-specific kinase 2
- Serine/threonine-protein kinase SRPK2
- SFRS protein kinase 2
- SFRSK2
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
SRPK
SubFamily:
NA
Structure
Mol. Mass (Da):
77,543
# Amino Acids:
688
# mRNA Isoforms:
2
mRNA Isoforms:
79,029 Da (699 AA; P78362-2); 77,527 Da (688 AA; P78362)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 81 | 686 | Pkinase |
| 331 | 374 | Coiled-coil |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S10, S52+, S250, S312, S380, S423, S472, S483, S484, S490, S494, S496, S497, S588+, S616.
Threonine phosphorylated:
T328, T376, T478, T492+, T498, T547.
Tyrosine phosphorylated:
Y182, Y319, Y425, Y427, Y431.
Ubiquitinated:
K179.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 20
20
957
64
1044
 1.4
1.4
65
28
40
 14
14
649
18
558
 16
16
771
202
925
 20
20
926
53
678
 5
5
232
165
842
 4
4
195
69
398
 25
25
1161
77
1979
 13
13
597
31
473
 3
3
122
159
244
 6
6
292
55
625
 15
15
704
324
656
 6
6
303
62
562
 1.4
1.4
65
21
40
 7
7
328
43
544
 2
2
71
35
62
 2
2
109
241
282
 8
8
368
33
511
 2
2
103
172
170
 14
14
667
238
676
 14
14
654
39
880
 6
6
288
47
474
 9
9
438
30
565
 35
35
1668
33
2030
 12
12
575
39
827
 18
18
841
124
924
 4
4
193
65
315
 8
8
390
33
545
 10
10
451
33
606
 3
3
124
56
100
 14
14
659
48
569
 54
54
2550
71
6355
 7
7
331
115
356
 17
17
816
135
701
 100
100
4711
83
6575
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.4
99.4
99.4
99 99.1
99.1
99.2
100 -
-
-
97 -
-
-
- 95.6
95.6
96.5
96 -
-
-
- 93.9
93.9
95.4
95 -
-
-
93 -
-
-
- 87.8
87.8
91.9
- -
-
-
91 80.7
80.7
87.4
85.5 -
-
-
78 -
-
-
- -
-
-
46 -
-
-
- 23.7
23.7
36
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
36 22.6
22.6
37.2
35 -
-
-
44
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | LUC7L3 - O95232 |
| 2 | SFRS6 - Q13247 |
| 3 | SNRNP70 - P08621 |
| 4 | RNPS1 - Q15287 |
| 5 | PPIL3 - Q9H2H8 |
| 6 | CLK1 - P49759 |
Regulation
Activation:
Activated by phosphorylation at Ser-52 and Ser-588.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
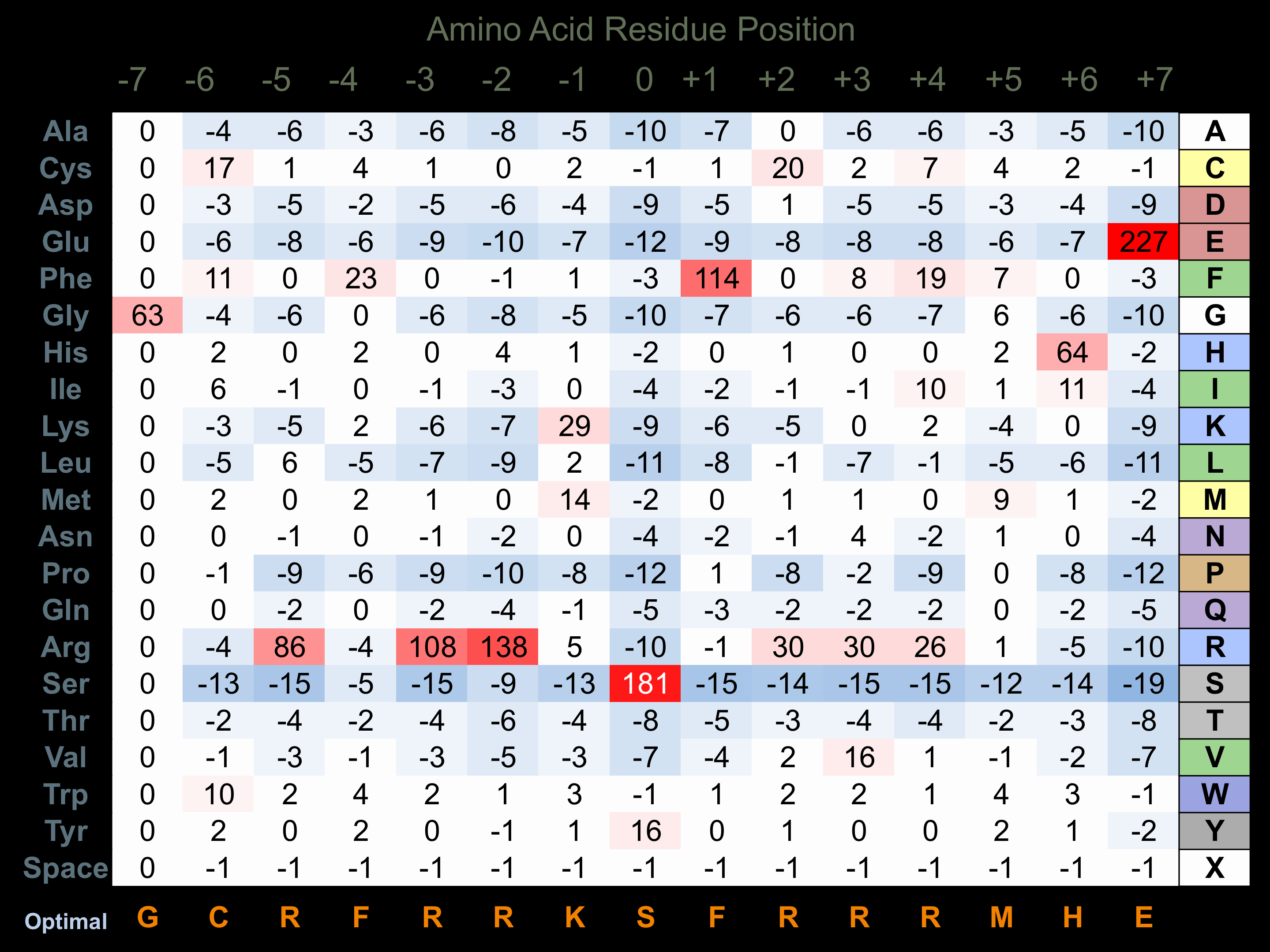
Matrix Type:
Experimentally derived from alignment of 1 known protein substrate phosphosites and 24 peptides phosphorylated by recombinant SRPK2 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Neurological disorder
Specific Diseases (Non-cancerous):
Alzheimer's disease (AD)
Comments:
SRPK2 has been shown to phosphorylate tau at S214, and contribute to the progression of Alzheimer's disease. It has also been noted to phosphorylate hepatitis B core protein.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +57, p<0.003); Cervical cancer stage 2A (%CFC= +52, p<0.032); Ovary adenocarcinomas (%CFC= +96, p<0.049); and Skin squamous cell carcinomas (%CFC= +77, p<0.066).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.04 % in 24442 diverse cancer specimens. This rate is -42 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.35 % in 589 stomach cancers tested; 0.33 % in 1093 large intestine cancers tested; 0.15 % in 1226 kidney cancers tested; 0.08 % in 1983 central nervous system cancers tested.
Frequency of Mutated Sites:
None > 5 in 20,056 cancer specimens
Comments:
Only 1 deletion, 3 insertions and no complex mutations are noted on the COSMIC website.

