Nomenclature
Short Name:
DNAPK
Full Name:
DNA-dependent protein kinase catalytic subunit
Alias:
- DNA-PK catalytic subunit
- DNA-PKcs
- PRKDC
- XRCC7
- DNPK1
- EC 2.7.11.1
- P460
- PRKD
Classification
Type:
Protein-serine/threonine kinase
Group:
Atypical
Family:
PIKK
SubFamily:
DNAPK
Specific Links
Structure
Mol. Mass (Da):
469,089
# Amino Acids:
4128
# mRNA Isoforms:
2
mRNA Isoforms:
469,089 Da (4128 AA; P78527); 465,501 Da (4097 AA; P78527-2)
4D Structure:
DNA-PK is a heterotrimer of PRKDC and the Ku p70-p86 (XRCC6-XRCC5) dimer. Formation of this complex may be promoted by interaction with ILF3. Associates with the DNA-bound Ku heterodimer, but it can also bind to and be activated by free DNA. Interacts with DNA-PKcs-interacting protein (KIP) with the region upstream the kinase domain. PRKDC alone also interacts with and phosphorylates DCLRE1C, thereby activating the latent endonuclease activity of this protein. Interacts with C1D. Interacts with TTI1 and TELO2
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
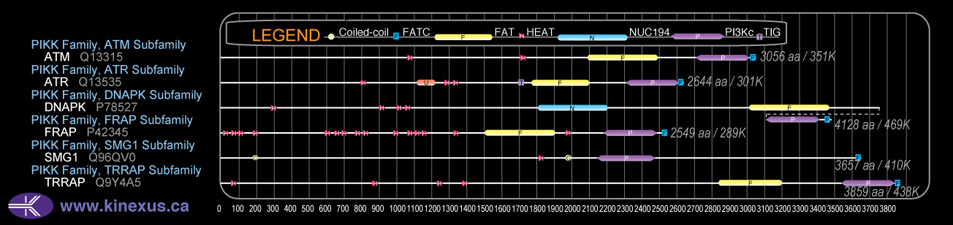
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
A2, K117 (N6), K254 (N6), K263,K357, K717, K810, K828, K832 (N6), K1057 (N6), K1074, K1209 (N6), K1970 (N6), K1985, K2259 (N6), K2359, K2445, K2683, K2694, K2702, K2738, K3239, K3241 (N6), K3260 (N6), K3267, K3608 (N6), K3621 (N6), K3638 (N6), K3642 (N6)K3691, K4048.
Methylated:
R820, K2702, K3372.
Serine phosphorylated:
S210, S318, S492, S503, S505, S511, S552, S687, S841, S842, S847, S861, S875, S876, S882, S893, S922, S1052, S1065, S1187, S1470, S1545, S1546, S1861, S2023, S2029, S2034, S2041, S2051, S2053, S2056, S2107, S2117, S2250, S2599, S2612-, S2624, S2626, S2654, S2655, S2672, S2674, S2675, S2677, S2695, S2789, S2814, S2932, S2955, S2998, S3010, S3018, S3021, S3205, S3215, S3233, S3253, S3290, S3294, S3363, S3366, S3432, S3489, S3497, S3500, S3546, S3657, S3731, S3821, S3870, S3993, S4026, S4096, .
Threonine phosphorylated:
T137, T516, T1275, T1481, T1620, T1621, T1856, T2535, T2600, T2603, T2609+, T2615, T2618, T2620, T2638-, T2645, T2647-, T2671, T3198, T3663, T3811, T3950-.
Tyrosine phosphorylated:
Y366, Y667, Y682, Y721, Y779, Y883+, Y1064, Y1086, Y2329, Y2440, Y2546, Y2743, Y2936, Y3378, Y3791, Y3828, Y3855, Y3859.
Ubiquitinated:
K99, K108, K117, K148, K155, K163, K164, K205, K225, K236, K254, K263, K310, K321, K329, K337, K357, K374, K379, K393, K471, K493, K525, K574, K681, K689, K692, K712, K801, K810, K824, K828, K838, K868, K881, K902, K963, K1038, K1042, K1051, K1057, K1074, K1141, K1147, K1186, K1193, K1209, K1311, K1334, K1357, K1407, K1412, K1422, K1456, K1489, K1612, K1627, K1642, K1857, K1869, K1870, K1875, K1892, K1895, K1913, K1917, K1970, K1973, K1985, K2003, K2127, K2132, K2259, K2263, K2334, K2347, K2350, K2366, K2369, K2418, K2433, K2447, K2503, K2549, K2683, K2694, K2702, K2703, K2715, K2738, K2752, K2758, K2764, K2786, K2806, K2829, K2835, K2908, K2928, K2950, K2970, K2978, K3050, K3067, K3075, K3128, K3147, K3158, K3172, K3192, K3196, K3235, K3241, K3248, K3257, K3260, K3264, K3302, K3318, K3355, K3372, K3426, K3449, K3452, K3455, K3552, K3598, K3621, K3631, K3638, K3669, K3691, K3710, K3753, K3825, K3840, K3845, K3860, K3884, K4007, K4019, K4023, K4036, K4050, K4070 .
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 33
33
867
48
1091
 3
3
80
24
71
 5
5
134
67
138
 17
17
457
191
877
 30
30
796
35
720
 5
5
129
117
167
 16
16
419
51
638
 44
44
1174
115
4318
 19
19
497
24
417
 4
4
97
193
113
 4
4
102
99
126
 18
18
473
342
546
 6
6
158
100
172
 3
3
92
21
71
 4
4
111
26
131
 3
3
83
24
68
 4
4
116
357
95
 5
5
121
82
113
 4
4
116
195
107
 22
22
589
162
675
 5
5
145
89
143
 5
5
143
94
140
 5
5
143
86
152
 5
5
146
83
154
 7
7
198
89
211
 50
50
1333
138
4460
 6
6
152
103
157
 4
4
103
83
108
 6
6
153
82
162
 2
2
51
42
59
 28
28
744
30
739
 100
100
2667
56
7600
 16
16
420
89
796
 26
26
691
88
630
 4
4
99
48
130
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.5
99.5
99.9
99.5 97.6
97.6
98.8
98 -
-
-
78 -
-
-
- -
-
-
83 -
-
-
- 79.2
79.2
88.8
79 39.2
39.2
44.7
79 -
-
-
- 74.2
74.2
85.4
- 68.8
68.8
82.6
69 64
64
79.8
64 58.5
58.5
76.2
59 -
-
-
- -
-
-
- -
-
-
- -
-
-
- 20.8
20.8
31.6
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | XRCC5 - P13010 |
| 2 | TP53 - P04637 |
| 3 | RPA2 - P15927 |
| 4 | XRCC6 - P12956 |
| 5 | CIB1 - Q99828 |
| 6 | DCLRE1C - Q96SD1 |
| 7 | NCOA6 - Q14686 |
| 8 | WRN - Q14191 |
| 9 | CHEK1 - O14757 |
| 10 | C1D - Q13901 |
| 11 | AKT1 - P31749 |
| 12 | HSP90AA1 - P07900 |
| 13 | PRKCD - Q05655 |
| 14 | PARP1 - P09874 |
| 15 | CASP3 - P42574 |
Regulation
Activation:
NA
Inhibition:
Phosphorylation at Ser-612, Thr-2609, Thr-2638, Thr-2647 and Thr-3950 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| AurB | Q96GD4 | S511 | ESEDHRASGEVRTGK | |
| DNAPK | P78527 | S2056 | VQSYSYSSQDPRPAT | ? |
| ATR | Q13535 | T2609 | LTPMFVETQASQGTL | + |
| ATM | Q13315 | T2609 | LTPMFVETQASQGTL | + |
| DNAPK | P78527 | T2609 | LTPMFVETQASQGTL | + |
| ATR | Q13535 | S2612 | MFVETQASQGTLQTR | ? |
| ATM | Q13315 | S2612 | MFVETQASQGTLQTR | ? |
| DNAPK | P78527 | S2612 | MFVETQASQGTLQTR | ? |
| ATM | Q13315 | T2638 | VAGQIRATQQQHDFT | - |
| DNAPK | P78527 | T2638 | VAGQIRATQQQHDFT | - |
| ATM | Q13315 | T2647 | QQHDFTLTQTADGRS | - |
| ATR | Q13535 | T2647 | QQHDFTLTQTADGRS | - |
| DNAPK | P78527 | T2647 | QQHDFTLTQTADGRS | - |
| DNAPK | P78527 | T3950 | GHAFGSATQFLPVPE | - |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| AIRE | O43918 | S156 | RGTASPGSQLKAKPP | |
| AIRE | O43918 | T68 | ALLSWLLTQDSTAIL | |
| Akt1 (PKBa) | P31749 | S473 | RPHFPQFSYSASGTA | + |
| Artemis | Q96SD1 | S503 | NDEITDESLENFPSS | |
| Artemis | Q96SD1 | S516 | SSTVAGGSQSPKLFS | |
| Artemis | Q96SD1 | S534 | GESTHISSQNSSQST | |
| Artemis | Q96SD1 | S538 | HISSQNSSQSTHITE | |
| Artemis | Q96SD1 | S548 | THITEQGSQGWDSQS | |
| Artemis | Q96SD1 | S553 | QGSQGWDSQSDTVLL | |
| Artemis | Q96SD1 | S645 | NLSTNADSQSSSDFE | |
| Caspase 2 | P42575 | S139 | LSCDYDLSLPFPVCE | |
| Chk2 (CHEK2) | O96017 | T383 | GETSLMRTLCGTPTY | + |
| Chk2 (CHEK2) | O96017 | T387 | LMRTLCGTPTYLAPE | - |
| Chk2 (CHEK2) | O96017 | T68 | SSLETVSTQELYSIP | + |
| DNAPK (PRKDC) | P78527 | S2056 | VQSYSYSSQDPRPAT | ? |
| DNAPK (PRKDC) | P78527 | S2612 | MFVETQASQGTLQTR | ? |
| DNAPK (PRKDC) | P78527 | T2609 | LTPMFVETQASQGTL | + |
| DNAPK (PRKDC) | P78527 | T2638 | VAGQIRATQQQHDFT | - |
| DNAPK (PRKDC) | P78527 | T2647 | QQHDFTLTQTADGRS | - |
| DNAPK (PRKDC) | P78527 | T3950 | GHAFGSATQFLPVPE | - |
| GR | P04150 | S508 | QQATTGVSQETSENP | |
| H2AX/H2AFX | P16104 | S139 | GKKATQASQEY____ | ? |
| hnRNP P2 | P35637 | S131 | QPQSGSYSQQPSYGG | |
| hnRNP P2 | P35637 | S26 | TQPGQGYSQQSSQPY | |
| hnRNP P2 | P35637 | S42 | QQSYSGYSQSTDTSG | |
| hnRNP P2 | P35637 | S61 | SYSSYGQSQNTGYGT | |
| hnRNP P2 | P35637 | S84 | STGGYGSSQSSQSSY | |
| hnRNP U | Q00839 | S59 | AMEPGNGSLDLGGDS | |
| HSP90AA1 | P07900 | T5 | ____PEETQTQDQPM | |
| HSP90AA1 | P07900 | T7 | __PEETQTQDQPMEE | |
| IRF3 | Q14653 | T135 | GGGSTSDTQEDILDE | + |
| Ku70 | P12956 | S5 | ___SGWESYYKTEGD | |
| Ku80 | P13010 | S576 | EQGGAHFSVSSLAEG | |
| Ku80 | P13010 | S579 | GAHFSVSSLAEGSVT | |
| Ku80 | P13010 | T714 | KDKPSGDTAAVFEEG | |
| LIG4 | P49917 | T650 | HLKAPNLTNVNKISN | |
| LKB1 (STK11) | Q15831 | S428 | SSKIRRLSACKQQ__ | + |
| LKB1 (STK11) | Q15831 | T363 | IEDGIIYTQDFTVPG | + |
| NFkB-p105 | P19838 | S20 | QMFHLDPSLTHTIFN | + |
| NHEJ1 | Q9H9Q4 | S132 | LASPSLVSQHLIRPL | |
| NHEJ1 | Q9H9Q4 | S245 | PHTSNSASLQGIDSQ | |
| NHEJ1 | Q9H9Q4 | S251 | ASLQGIDSQCVNQPE | |
| NHEJ1 | Q9H9Q4 | S263 | QPEQLVSSAPTLSAP | |
| NHEJ1 | Q9H9Q4 | S55 | QVDTSVVSQRAKELN | |
| NHEJ1 | Q9H9Q4 | T223 | DLYMAVTTQEVQVGQ | |
| NHEJ1 | Q9H9Q4 | T266 | QLVSSAPTLSAPEKE | |
| p53 | P04637 | S15 | PSVEPPLSQETFSDL | + |
| p53 | P04637 | S20 | PLSQETFSDLWKLLP | + |
| p53 | P04637 | S37 | NVLSPLPSQAMDDLM | + |
| p53 | P04637 | S46 | AMDDLMLSPDDIEQW | + |
| p53 | P04637 | S9 | EEPQSDPSVEPPLSQ | |
| p53 | P04637 | T18 | EPPLSQETFSDLWKL | + |
| PDX1 | P52945 | T11 | EEQYYAATQLYKDPC | - |
| POU2F1 | P14859 | S141 | AAAVQQHSASQQHSA | - |
| POU2F1 | P14859 | S143 | AVQQHSASQQHSAAG | - |
| POU2F1 | P14859 | S147 | HSASQQHSAAGATIS | - |
| POU2F1 | P14859 | S167 | PMTQIPLSQPIQIAQ | - |
| POU2F1 | P14859 | S232 | LTQLPQQSQANLLQS | - |
| POU2F1 | P14859 | S78 | QSKSNEESGDSQQPS | - |
| POU2F1 | P14859 | S81 | SNEESGDSQQPSQPS | - |
| POU2F1 | P14859 | S85 | SGDSQQPSQPSQQPS | - |
| POU2F1 | P14859 | S88 | SQQPSQPSQQPSVQA | |
| POU2F1 | P14859 | S92 | SQPSQQPSVQAAIPQ | - |
| POU2F1 | P14859 | T100 | VQAAIPQTQLMLAGG | - |
| POU2F1 | P14859 | T162 | ASAATPMTQIPLSQP | - |
| POU2F1 | P14859 | T226 | LQAQNLLTQLPQQSQ | - |
| RPA2 | P15927 | S33 | GFGSPAPSQAEKKSR | |
| RPA2 | P15927 | S4 | ____MWNSGFESYGS | |
| RPA2 | P15927 | S8 | MWNSGFESYGSSSYG | |
| RPA2 | P15927 | T21 | YGGAGGYTQSPGGFG | |
| SRF | P11831 | S435 | LTVLNAFSQAPSTMQ | |
| SRF | P11831 | S446 | STMQVSHSQVQEPGG | |
| USF1 | P22415 | S262 | RQSNHRLSEELQGLD | |
| XPA | P23025 | S173 | VKKNPHHSQWGDMKL | |
| XPA | P23025 | S196 | RSLEVWGSQEALEEA | |
| XRCC1 | P18887 | S371 | FANTPKYSQVLGLGG | |
| XRCC4 | Q13426 | S260 | KDDSIISSLDVTDIA | |
| XRCC4 | Q13426 | S304 | ENSRPDSSLPETSKK | |
| XRCC4 | Q13426 | S320 | HISAENMSLETLRNS | |
| XRCC4 | Q13426 | S327 | SLETLRNSSPEDLFD | |
| XRCC4 | Q13426 | S328 | LETLRNSSPEDLFDE | |
| Zic2 | O95409 | S199 | EQYRQVASPRTDPYS |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
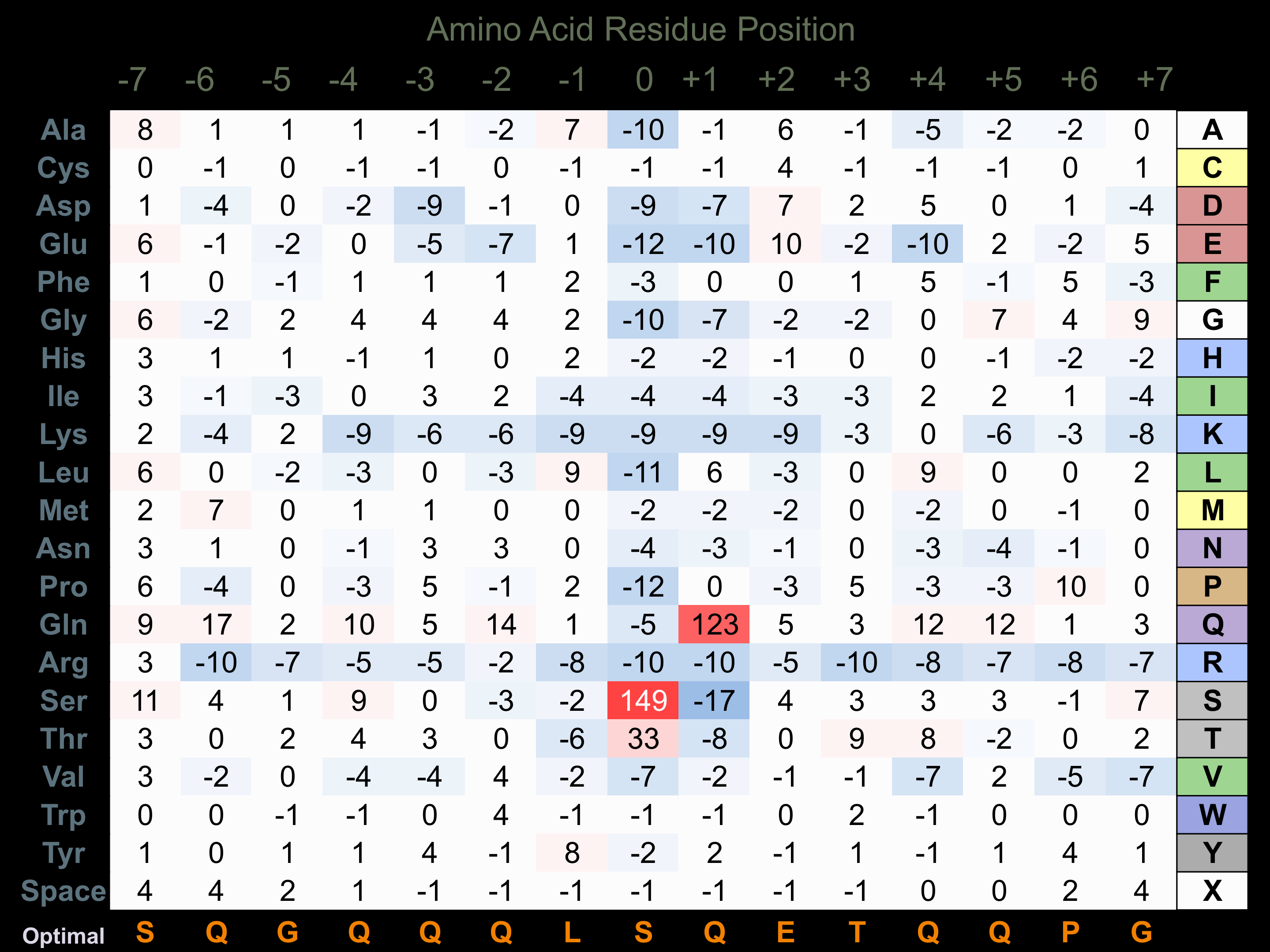
Matrix Type:
Experimentally derived from alignment of 101 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| PI-103 | IC50 = 2 nM | 16739368 | 538346 | |
| Torin1 | IC50 = 6.34 nM | 49836027 | 1255226 | |
| PP121 | IC50 = 60 nM | 24905142 | 18849971 | |
| DNA-PK Inhibitor III | IC50 = 120 nM | 9859309 | 1317546 | 14707266 |
| Wortmannin | Ki = 120 nM | 312145 | 428496 | 15658870 |
| XL765 | IC50 = 150 nM | 49867926 | ||
| LY293646 | IC50 = 230 nM | 9860529 | 104468 | 12941339 |
| AMA37 | IC50 = 270 nM | 16760391 | 478980 | 17562705 |
| PP242 | IC50 = 410 nM | 25243800 | 18849971 | |
| LY294002 | IC50 = 1.2 µM | 3973 | 98350 | 15546735 |
| XL147 | IC50 = 4.75 µM | 1893730 |
Disease Linkage
General Disease Association:
Immune disorders
Specific Diseases (Non-cancerous):
Artemis deficiency; Severe combined immunodeficiency due to DNA-PKCS deficiency (SCID)
Comments:
Artemis Deficiency is linked to inhibition to DNA repair mechanisms and can be associated with inflammatory bowel disease. Severe Combined Immunodeficiency Due to Dna-Pkcs Deficiency is an immunodeficiency disorder. C1D to DNAPK interaction can be inhibited with the L1510P, or the E1516P + L1517D mutations. Radiation sensitivity and partially inhibited binding of DSB to DNAPK occurs with the mutation T2609A, but only when in association with the reduction of phosphorylation mutation S2612A mutation. If the mutations T2638A and T2647A occur in conjunction then phosphorylation is reduced, and DNAPK is constitutively active but would also be sensitive to radioactivity.
Comments:
DNAPK appears to be a tumour suppressor protein (TSP). The active form of the protein kinase normally acts to inhibit tumour cell proliferation until damage to DNA can be repaired. Therefore, it can play a role to facilitate tumour cell survival under conditions of radiation and chemotherapy.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +152, p<0.0002);Cervical cancer (%CFC= -54, p<0.0001); Cervical cancer stage 2B (%CFC= -62, p<0.089); Colon mucosal cell adenomas (%CFC= +71, p<0.0001); Gastric cancer (%CFC= +219, p<0.0001); Lung adenocarcinomas (%CFC= +78, p<0.0001); Malignant pleural mesotheliomas (MPM) tumours (%CFC= +50, p<0.043); Oral squamous cell carcinomas (OSCC) (%CFC= +124, p<0.0001); Skin fibrosarcomas (%CFC= +138, p<0.066); Skin melanomas - malignant (%CFC= +115, p<0.0001); Skin squamous cell carcinomas (%CFC= +84, p<0.09); T-cell prolymphocytic leukemia (%CFC= +110, p<0.035); and Vulvar intraepithelial neoplasia (%CFC= +116, p<0.011).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 24904 diverse cancer specimens. This rate is only -26 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.29 % in 1052 large intestine cancers tested; 0.19 % in 555 stomach cancers tested; 0.14 % in 602 endometrium cancers tested; 0.13 % in 805 skin cancers tested; 0.1 % in 1727 lung cancers tested; 0.09 % in 814 upper aerodigestive tract cancers tested; 0.07 % in 605 oesophagus cancers tested; 0.07 % in 500 urinary tract cancers tested; 0.05 % in 1270 liver cancers tested; 0.04 % in 1439 breast cancers tested; 0.03 % in 878 ovary cancers tested; 0.03 % in 1203 kidney cancers tested; 0.03 % in 1173 pancreas cancers tested.
Frequency of Mutated Sites:
None > 7 in 20,763 cancer specimens
Comments:
Over 26 deletions, 10 insertions and 2 complex mutations are noted on the COSMIC website.

