Nomenclature
Short Name:
PCTAIRE1
Full Name:
Serine-threonine-protein kinase PCTAIRE-1
Alias:
- CDK16
- PCTAIRE-motif protein kinase 1
- PCTGAIRE
- PCTK1
- Serine/threonine-protein kinase PCTAIRE1
- EC 2.7.11.22
- FLJ16665
- KPT1
- PCTAIRE
- PCTAIRE protein kinase 1
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
CDK
SubFamily:
TAIRE
Specific Links
Structure
Mol. Mass (Da):
55,716
# Amino Acids:
496
# mRNA Isoforms:
3
mRNA Isoforms:
63,458 Da (570 AA; Q00536-2); 56,375 Da (502 AA; Q00536-3); 55,716 Da (496 AA; Q00536)
4D Structure:
Found in a complex containing CABLES1, CDK17 and TDRD7. Interacts with YWHAH, YWHAQ and YWHAZ
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 165 | 446 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K108, K116.
Serine phosphorylated:
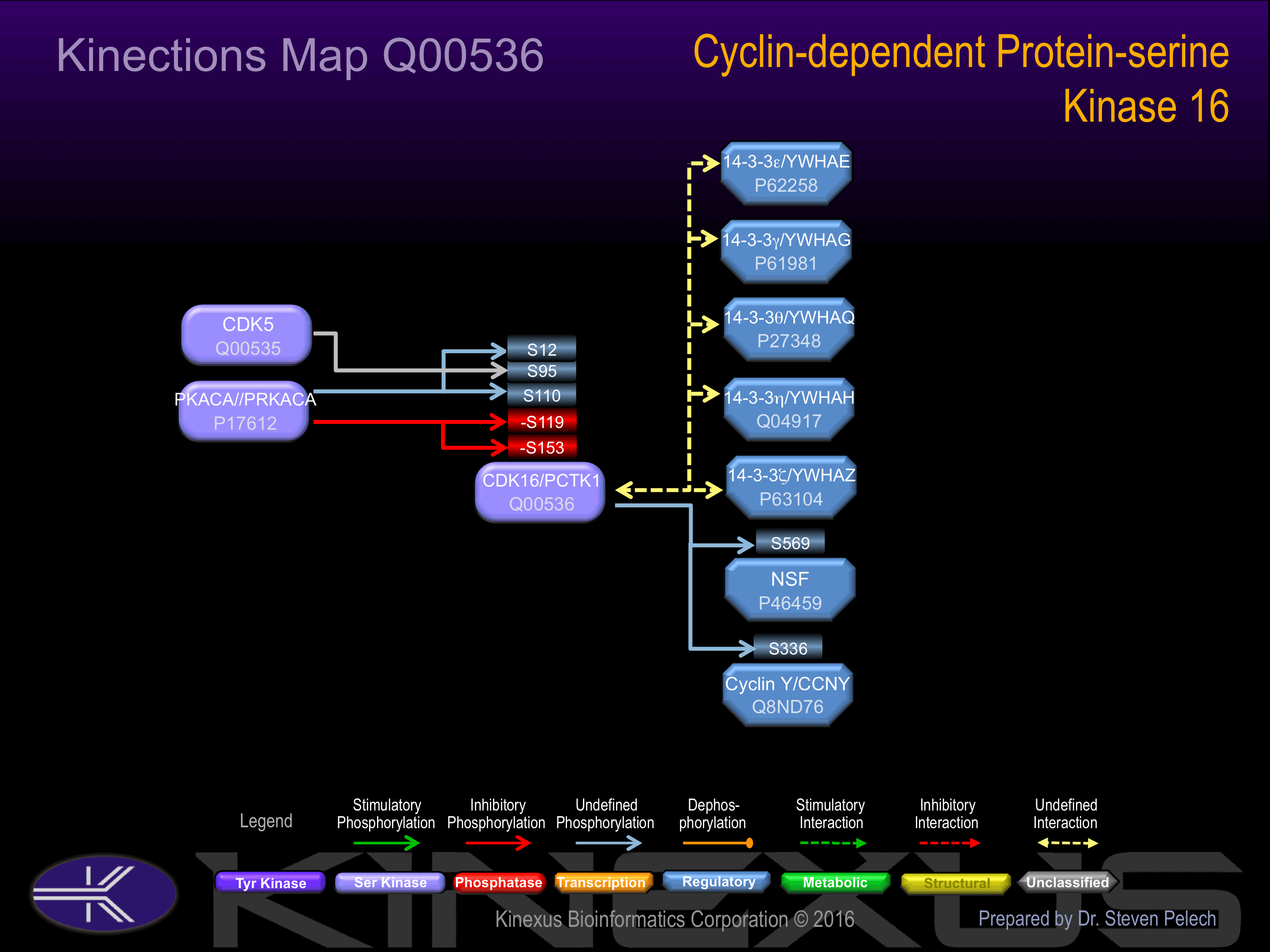
S12, S36, S42, S64, S65, S78, S82, S86, S89, S95, S110, S119-, S138, S153-, S155+, S319+, S479, S480.
Threonine phosphorylated:
T14, T88, T111, T135, T175, T178.
Tyrosine phosphorylated:
Y176, Y180.
Ubiquitinated:
K133, K143, K167, K194, K301, K455, K466.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 65
65
1010
39
975
 12
12
187
24
79
 17
17
272
33
161
 34
34
523
162
491
 55
55
853
36
666
 8
8
117
119
131
 25
25
384
51
565
 59
59
920
83
1033
 33
33
514
24
332
 17
17
265
159
205
 9
9
139
64
123
 42
42
661
286
647
 13
13
198
66
184
 7
7
113
20
56
 18
18
275
57
303
 10
10
160
24
91
 15
15
239
326
1805
 22
22
340
48
369
 31
31
485
167
383
 33
33
516
162
511
 20
20
309
54
265
 13
13
200
60
183
 16
16
253
44
180
 22
22
336
48
220
 14
14
217
53
195
 54
54
840
102
1057
 14
14
218
69
171
 17
17
261
48
202
 16
16
250
47
202
 19
19
289
42
117
 24
24
380
42
296
 100
100
1557
51
2841
 33
33
514
89
942
 50
50
786
83
686
 16
16
251
48
256
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 58
58
60.8
100 89.3
89.3
90.3
99.5 -
-
-
98 -
-
-
91 97.5
97.5
98
92 -
-
-
- 97.5
97.5
98.9
98 95.5
95.5
97.3
98 -
-
-
- 76.1
76.1
81
- 69.1
69.1
82.9
- 45.1
45.1
59.6
- 41.9
41.9
55.8
80.5 -
-
-
- 35.4
35.4
43.9
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 33
33
45.3
- -
-
-
- 33.4
33.4
46.3
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | YWHAG - P61981 |
| 2 | CDK5 - Q00535 |
| 3 | YWHAZ - P63104 |
| 4 | YWHAH - Q04917 |
| 5 | CDK5R1 - Q15078 |
| 6 | SEC23A - Q15436 |
| 7 | PRKACA - P17612 |
| 8 | YWHAE - P62258 |
| 9 | CCNB2 - O95067 |
| 10 | CNNY-Q8ND76 |
Regulation
Activation:
Phosphorylation at Ser-119 induces interaction with 14-3-3 gamma and 14-3-3 zeta.
Inhibition:
Phosphorylation at Ser-153 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
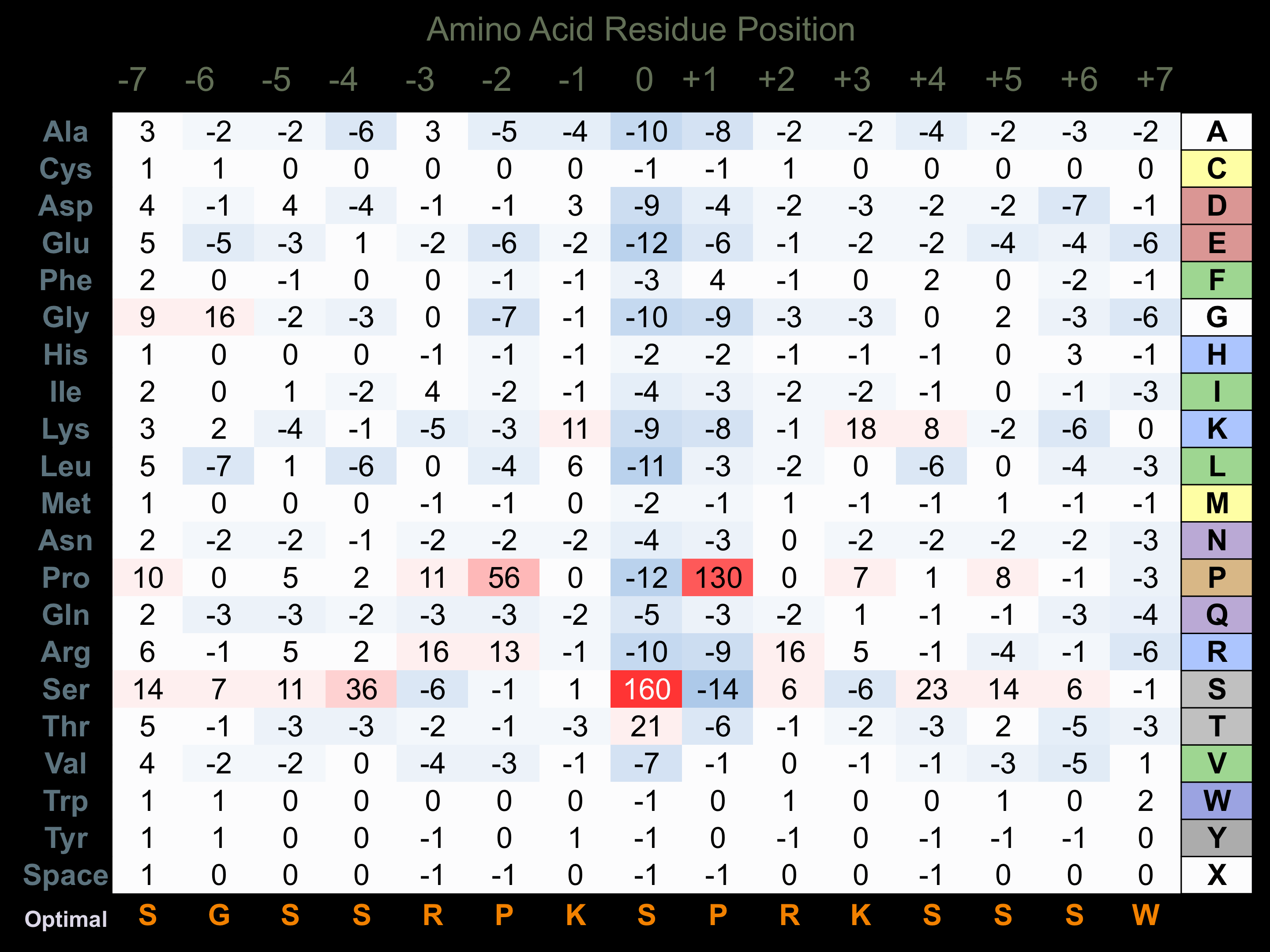
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= +231, p<0.005); Brain oligodendrogliomas (%CFC= +382, p<0.013); Cervical cancer stage 2A (%CFC= +131, p<0.029); Cervical cancer stage 2B (%CFC= +146, p<0.097); Lung adenocarcinomas (%CFC= +109, p<0.0001); Ovary adenocarcinomas (%CFC= +97, p<0.0008); Prostate cancer - metastatic (%CFC= +77, p<0.0001); Skin fibrosarcomas (%CFC= +182, p<0.002); Skin melanomas - malignant (%CFC= +167, p<0.0001). The COSMIC website notes an up-regulated expression score for PCTAIRE1 in diverse human cancers of 573, which is 1.2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 71 for this protein kinase in human cancers was 1.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis. BRSK2 mediated phosphorylation of PCTAIRE1 can be inhibited with S12A mutation, which leads to inhibition of insulin secretion. The interaction of PCTAIRE1 and the cyclin CCNY can be inhibited with a S119A mutation. PCTAIRE1 phosphotransferase activity can be constitutively activated through the mutation S153A, leading to increased interaction of PCTAIRE1 and CCNY. Interaction with CCNY can be inhibited with a S153D mutation. The phosphotransferase activity of the PCTAIRE 1 can be inhibited with a K194A mutation, leading to inhibited effect of PCTAIRE1 on insulin secretion. PCTAIRE1 phosphotransferase activity is lost with the K194R mutation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.04 % in 24939 diverse cancer specimens. This rate is -41 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Frequency of Mutated Sites:
None > 4 in 20,222 cancer specimens
Comments:
Only 1 insertion, and no deletions or complex mutations are noted on the COSMIC website.