Nomenclature
Short Name:
ACK1
Full Name:
Activated CDC42 kinase 1
Alias:
- ACK
- Kinase ACK1
- Non-receptor protein tyrosine kinase Ack
- P21cdc42Hs
- TNK2
- Tyrosine kinase, non-receptor, 2
- Activated Cdc42-associated kinase 1
- Activated p21cdc42Hs kinase
- Activated p21cdc42Hs kinase 1
- EC 2.7.10.2
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Ack
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
114,569
# Amino Acids:
1038
# mRNA Isoforms:
3
mRNA Isoforms:
119,349 Da (1086 AA; Q07912-3); 114,569 Da (1038 AA; Q07912); 60,062 Da (528 AA; Q07912-2)
4D Structure:
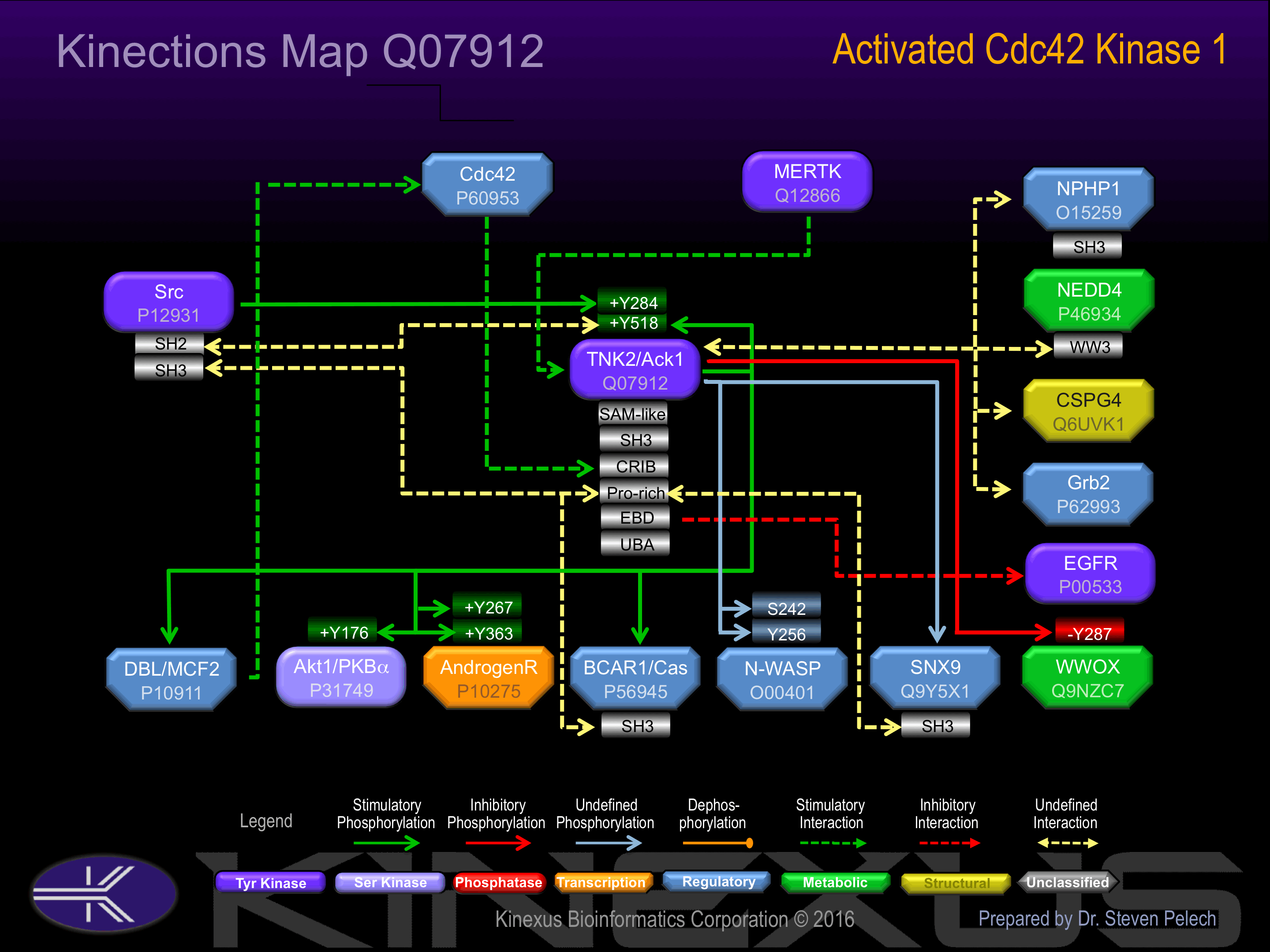
Interacts with CDC42. Interacts with activated CSPG4. Interacts with AR. Interacts with WWOX. Interacts with MERTK (activated); stimulates autophosphorylation. May interact (phosphorylated) with HSP90AB1; maintains kinase activity
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
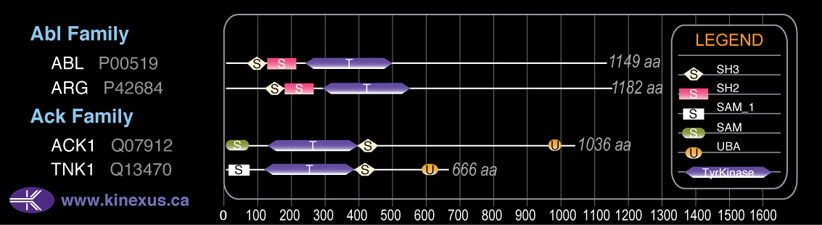
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K826.
Serine phosphorylated:
S36, S74, S102, S136, S149, S212, S607, S724, S728, S730, S785, S789, S810, S855, S856, S867, S881.
Threonine phosphorylated:
T101, T205, T465, T517, T603, T818, T829, T857, T925.
Tyrosine phosphorylated:
Y193, Y284+, Y518+, Y635, Y827, Y859, Y860, Y868, Y872.
Ubiquitinated:
K131.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
892
66
1179
 5
5
43
30
69
 8
8
69
15
58
 27
27
245
212
440
 63
63
558
61
498
 4
4
40
149
99
 27
27
243
84
571
 55
55
490
68
655
 35
35
315
27
314
 18
18
158
167
278
 9
9
76
50
103
 69
69
617
300
566
 9
9
84
48
114
 6
6
56
24
61
 18
18
162
42
241
 5
5
46
39
104
 33
33
291
321
2165
 10
10
92
33
118
 9
9
82
156
84
 43
43
384
239
381
 22
22
195
39
215
 16
16
139
45
190
 19
19
169
26
193
 22
22
192
33
729
 19
19
168
39
255
 95
95
848
136
1643
 6
6
55
54
75
 15
15
131
33
332
 8
8
75
33
94
 30
30
265
56
193
 86
86
767
42
768
 55
55
494
72
557
 5
5
44
148
184
 82
82
728
161
649
 11
11
96
87
68
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 26.5
26.5
34.5
0 98.5
98.5
98.8
99 -
-
-
95 -
-
-
93 94.3
94.3
95.8
95 -
-
-
- 92.9
92.9
94.2
95 93.8
93.8
95.6
94 -
-
-
- 38.3
38.3
45.9
- -
-
-
75 -
-
-
70 56.2
56.2
67.2
68 -
-
-
- 25.1
25.1
40.7
- 32.4
32.4
47.3
- 27.2
27.2
41.4
29 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | NCK1 - P16333 |
| 2 | SNX9 - Q9Y5X1 |
| 3 | CDC42 - P60953 |
| 4 | CLTC - Q00610 |
| 5 | EGFR - P00533 |
| 6 | HSH2D - Q96JZ2 |
| 7 | RASGRF1 - Q13972 |
| 8 | CSPG4 - Q6UVK1 |
| 9 | MERTK - Q12866 |
| 10 | ITFG2 - Q969R8 |
| 11 | SEZ6 - Q53EL9 |
Regulation
Activation:
Phosphorylation at Tyr-284 increases phosphotransferase activity.
Inhibition:
The SH3 domain appears to play an autoinhibitory role.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ACK1 (TNK2) | Q07912 | Y284 | LPQNDDHYVMQEHRK | + |
| ACK1 (TNK2) | Q07912 | Y518 | GGVKKPTYDPVSEDQ | + |
| Akt1 | P31749 | Y176 | EKATGRYYAMKILKK | + |
| AR (Androgen receptor) | P10275 | Y267 | QLRGDCMYAPLLGVP | + |
| AR (Androgen receptor) | P10275 | Y363 | AYQSRDYYNFPLALA | + |
| N-WASP (WAS-L) | O00401 | S242 | LFDMCGISEAQLKDR | |
| N-WASP (WAS-L) | O00401 | Y256 | RETSKVIYDFIEKTG | |
| WWOX | Q9NZC7 | Y287 | LSPTKNDYWAMLAYN | - |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
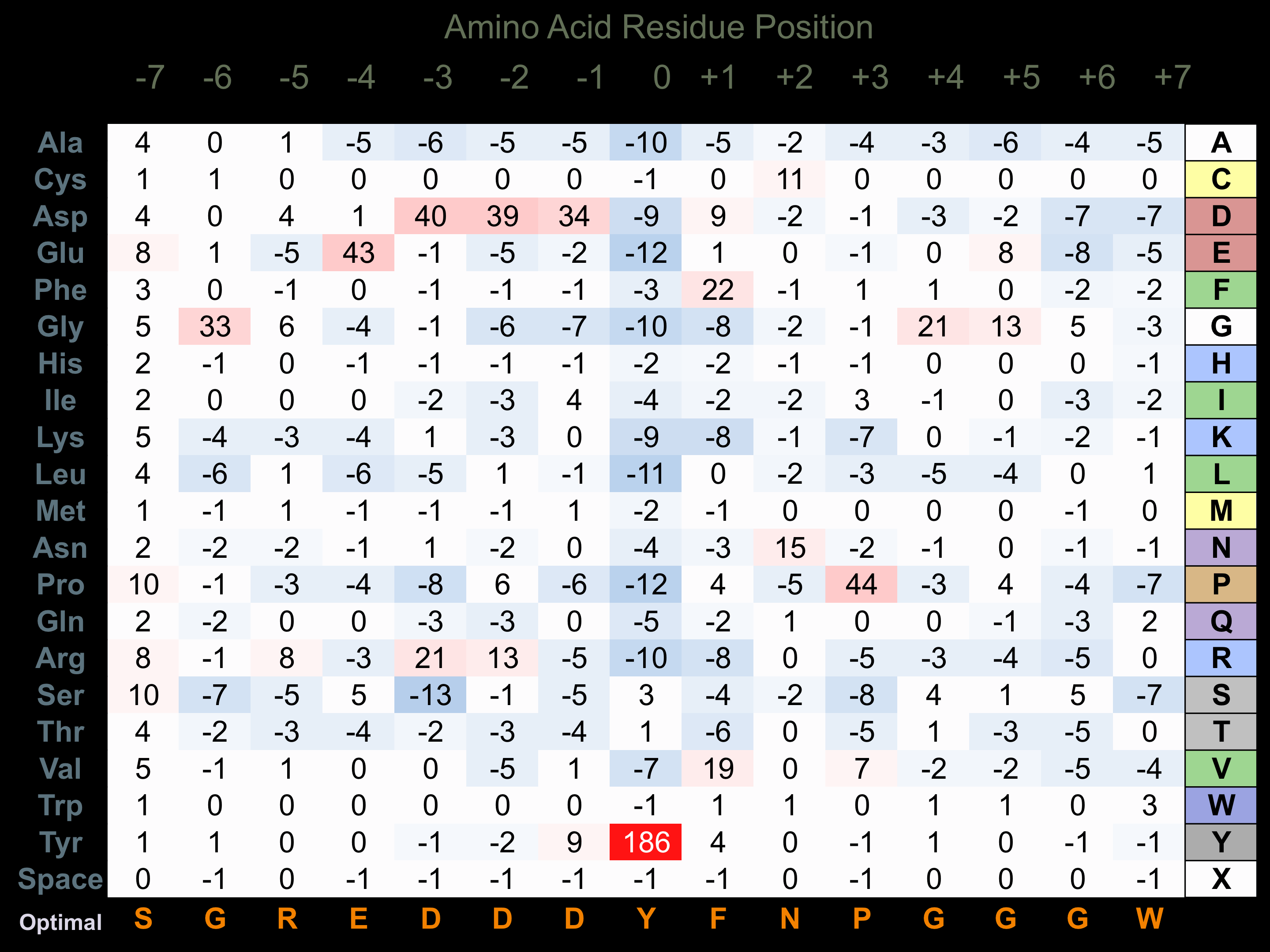
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, neurological disorders
Specific Diseases (Non-cancerous):
Infantile-Onset Mesial Temporal Lobe Epilepsy with Severe Cognitive Regression
Comments:
Infantile-Onset Mesial Temporal Lobe Epilepsy with Severe Cognitive Regression is a rare neuronal disease where ACK1 is implicated with a causative germline mutation (V716M). The mutated protein was expressed similarly to wildtype, but the the interaction of ACK1 with its ubiquitin ligases, is disrupted and this leads to loss of its degradation by EGF-mediated activation of cell signalling.
Specific Cancer Types:
Thyroid, Stomach and Eye
Comments:
ACK1 appears to be a tumour suppressor protein (TSP). Cancer-related mutations in human tumours point to a loss of function of the protein kinase. The active form of the protein kinase normally acts to inhibt tumour cell proliferation.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in human Uterine leiomyomas (%CFC= +65, p<0.066). The COSMIC website notes an up-regulated expression score for ACK1 in diverse human cancers of 709, which is 1.5-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 8 for this protein kinase in human cancers was 0.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24926 diverse cancer specimens. This rate is only -10 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.38 % in 1093 large intestine cancers tested; 0.35 % in 805 skin cancers tested; 0.18 % in 575 stomach cancers tested; 0.09 % in 1808 lung cancers tested; 0.06 % in 1962 central nervous system cancers tested.
Frequency of Mutated Sites:
None >5 in 20,190 cancer specimens