Nomenclature
Short Name:
RSK1
Full Name:
Ribosomal protein S6 kinase alpha 2
Alias:
- 90 kDa ribosomal protein S6 kinase 1
- EC 2.7.11.1
- MAPKAPK1A
- P90RSK1
- RPS6KA1
- S6K-alpha 1
- HU-1
- Kinase p90RSK1
- KS6A1
- KS6AA
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
RSK
SubFamily:
RSK
Specific Links
Structure
Mol. Mass (Da):
82,723
# Amino Acids:
735
# mRNA Isoforms:
4
mRNA Isoforms:
83,932 Da (744 AA; Q15418-2); 82,723 Da (735 AA; Q15418); 81,147 Da (719 AA; Q15418-4); 72,698 Da (643 AA; Q15418-3)
4D Structure:
Forms a complex with either ERK1 or ERK2 in quiescent cells. Transiently dissociates following mitogenic stimulation.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
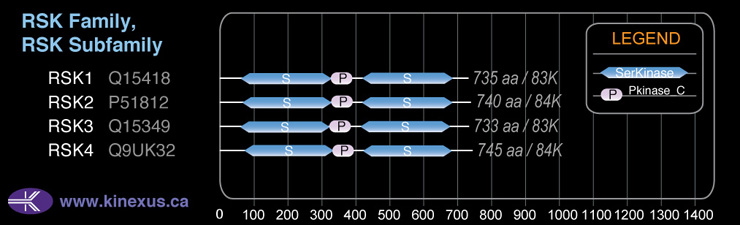
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K75.
Serine phosphorylated:
S45, S54, S72, S154-, S209+, S221+, S307, S363+, S369, S380+, S402, S637, S684, S687, S703, S732.
Threonine phosphorylated:
T225-, T359+, T384, T474, T522, T573+, T577-, T701, T733, T734.
Tyrosine phosphorylated:
Y91+, Y138, Y220+, Y228-, Y466, Y576+, Y702.
Ubiquitinated:
K66, K75, K278, K316, K500, K654.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 33
33
859
16
1191
 3
3
75
10
65
 1.5
1.5
38
1
0
 41
41
1054
48
2118
 30
30
783
14
752
 4
4
97
41
66
 20
20
518
19
677
 100
100
2581
25
4531
 29
29
759
10
683
 8
8
197
45
145
 3
3
82
14
74
 27
27
705
105
667
 6
6
147
12
33
 1
1
25
8
24
 4
4
101
9
92
 4
4
93
7
41
 25
25
647
101
3432
 3
3
75
7
46
 2
2
64
43
35
 21
21
542
56
550
 4
4
103
10
110
 14
14
362
12
313
 8
8
210
10
90
 1.3
1.3
34
8
45
 6
6
144
10
161
 59
59
1522
32
2706
 5
5
138
15
72
 3
3
70
8
54
 1.4
1.4
35
6
56
 3
3
85
14
69
 39
39
1005
30
710
 23
23
591
21
671
 7
7
191
56
303
 39
39
1014
31
719
 1.1
1.1
29
22
32
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
100 73
73
83.4
97 -
-
-
99 -
-
-
- 94.3
94.3
95.5
97 -
-
-
- 97
97
97.6
98.5 97.5
97.5
98.9
98 -
-
-
- 79.7
79.7
86.5
- 90
90
93.2
92 84.9
84.9
90.1
86 75.1
75.1
85.2
78 -
-
-
- 24.3
24.3
36.4
63 -
-
-
- 58.2
58.2
73
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | MAPK1 - P28482 |
| 2 | TSC2 - P49815 |
| 3 | MAPK3 - P27361 |
| 4 | CREB1 - P16220 |
| 5 | NFKBIA - P25963 |
| 6 | EEF2K - O00418 |
| 7 | BAD - Q92934 |
| 8 | EIF4B - P23588 |
| 9 | MITF - O75030 |
| 10 | PPP1R3A - Q16821 |
| 11 | TOB1 - P50616 |
| 12 | FOS - P01100 |
| 13 | ETV1 - P50549 |
| 14 | FGFR1 - P11362 |
| 15 | ESR1 - P03372 |
Regulation
Activation:
Phosphorylation at Ser-221 is required for increased phosphotransferase activity, but is insufficient by itself. Phosphorylation at Ser-363 and Ser-380 increases phosphotransferase activity. Phosphorylation at Ser-380 in Rsk2 permits recruitment and activation of PDK1 by autophosphorylation to then phosphorylate S227 (S221 in Rsk1). Phosphorylation of Thr-573 increases phosphotransferase activity.
Inhibition:
Phosphorylation at Ser-154 induces interaction with 14-3-3 and inhibits catalytic activity. Phosphorylation of Ser-732 permits dissociation of Rsk-ERK complex.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| RSK1 | Q15418 | S221 | DHEKKAYSFCGTVEY | + |
| PDK1 | O15530 | S221 | DHEKKAYSFCGTVEY | + |
| ERK1 | P27361 | T359 | DTEFTSRTPKDSPGI | + |
| ERK2 | P28482 | T359 | DTEFTSRTPKDSPGI | + |
| ERK2 | P28482 | S363 | TSRTPKDSPGIPPSA | + |
| PKCa | P17252 | S363 | TSRTPKDSPGIPPSA | + |
| ERK1 | P27361 | S363 | TSRTPKDSPGIPPSA | + |
| PDK1 | O15530 | S363 | TSRTPKDSPGIPPSA | + |
| RSK1 | Q15418 | S380 | HQLFRGFSFVATGLM | + |
| JNK2 | P45984 | S380 | HQLFRGFSFVATGLM | + |
| JNK1 | P45983 | S380 | HQLFRGFSFVATGLM | + |
| p70S6K | P23443 | S380 | HQLFRGFSFVATGLM | + |
| ERK2 | P28482 | T573 | AENGLLMTPCYTANF | + |
| ERK1 | P27361 | T573 | AENGLLMTPCYTANF | + |
| RSK1 | Q15418 | S732 | RRVRKLPSTTL____ |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Bad | Q92934 | S118 | GRELRRMSDEFVDSF | - |
| Bad | Q92934 | S75 | EIRSRHSSYPAGTED | - |
| Bad | Q92934 | S99 | PFRGRSRSAPPNLWA | - |
| C-EBPb | P17676 | T266 | VKSKAKKTVDKHSDE | |
| CaRHSP1 | Q9Y2V2 | S52 | TRRTRTFSATVRASQ | |
| CCT2 | P78371 | S260 | GSRVRVDSTAKVAEI | |
| CREB1 | P16220 | S133 | EILSRRPSYRKILND | + |
| DAPK1 | P53355 | S289 | QALSRKASAVNMEKF | - |
| DLC1 | Q96QB1 | S329 | VTRTRSLSACNKRVG | |
| eEF2K | O00418 | S366 | SPRVRTLSGSRPPLL | - |
| eIF4B | P23588 | S406 | RPRERHPSWRSEETQ | |
| eIF4B | P23588 | S409 | ERHPSWRSEETQERE | |
| eIF4B | P23588 | S422 | RERSRTGSESSQTGT | |
| ERa (ESR1) | P03372 | S167 | GGRERLASTNDKGSM | ? |
| ETV1 (ER81) | P50549 | S191 | HRFRRQLSEPCNSFP | + |
| ETV1 (ER81) | P50549 | S216 | PMYQRQMSEPNIPFP | + |
| ETV1 (ER81) | P50549 | S334 | PTYQRRGSLQLWQFL | |
| FLNA | P21333 | S2152 | TRRRRAPSVANVGSH | |
| Fos | P01100 | S362 | AAAHRKGSSSNEPSS | + |
| GMFb | P60983 | T26 | KFRFRKETNNAAIIM | |
| GSK3b | P49841 | S9 | SGRPRTTSFAESCKP | - |
| H3.1 | P68431 | S10 | TKQTARKSTGGKAPR | |
| H3.2 | P84228 | S11 | TKQTARKSTGGKAPR | |
| HMGN1 (HMG14) | P05114 | S6 | PKRKVSSAEGAAK__ | |
| IkBa | P25963 | S19 | DADEWCDSGLGSLGP | |
| IkBa | P25963 | S23 | WCDSGLGSLGPDAAA | |
| IkBa | P25963 | S32 | LLDDRHDSGLDSMKD | |
| IkBa | P25963 | S36 | RHDSGLDSMKDEEYE | |
| Kv4.3 | Q9UK17 | S535 | YPSTRSPSLSSHPGL | |
| Kv4.3 | Q9UK17 | S569 | LPATRLRSMQELSTI | |
| LKB1 (STK11) | Q15831 | S428 | SSKIRRLSACKQQ__ | + |
| Mad1 (MXD1) | Q05195 | S145 | IERIRMDSIGSTVSS | - |
| MDM2 | Q00987 | S166 | SSRRRAISETEENSD | |
| MDM2 | Q00987 | S186 | RQRKRHKSDSISLSF | |
| METTL1 | Q9UBP6 | S27 | YYRQRAHSNPMADHT | - |
| MITF | O75030 | S516 | KTSSRRSSMSMEETE | - |
| N-CAM L1 | P32004 | S1152 | RSKGGKYSVKDKEDT | |
| NDRG2 | Q9UN36 | S332 | LSRSRTASLTSAASV | |
| NDRG2 | Q9UN36 | S350 | RSRSRTLSQSSESGT | |
| nNOS | P29475 | S852 | SYKVRFNSVSSYSDS | |
| Nur77 | P22736 | S351 | GRRGRLPSKPKQPPD | - |
| p27Kip1 | P46527 | S10 | NVRVSNGSPSLERMD | - |
| p27Kip1 | P46527 | T198 | PGLRRRQT_______ | - |
| RANBP3 | Q9H6Z4 | S126 | VKRERTSSLTQFPPS | |
| Raptor | Q8N122 | S719 | PCTPRLRSVSSYGNI | |
| Raptor | Q8N122 | S721 | TPRLRSVSSYGNIRA | |
| Raptor | Q8N122 | S722 | PRLRSVSSYGNIRAV | |
| RPS6 | P62753 | S235 | IAKRRRLSSLRASTS | + |
| RPS6 | P62753 | S236 | AKRRRLSSLRASTSK | + |
| RSK1 (RPS6KA1) | Q15418 | S221 | DHEKKAYSFCGTVEY | + |
| RSK1 (RPS6KA1) | Q15418 | S380 | HQLFRGFSFVATGLM | + |
| RSK1 (RPS6KA1) | Q15418 | S732 | RRVRKLPSTTL____ | |
| SLC9A1 (NHE1) | P19634 | S703 | MSRARIGSDPLAYEP | + |
| SRF | P11831 | S103 | RGLKRSLSEMEIGMV | |
| TNNI3 | P19429 | S22 | PAPIRRRSSNYRAYA | |
| TNNI3 | P19429 | S23 | APIRRRSSNYRAYAT | |
| TSC2 | P49815 | S1798 | GQRKRLISSVEDFTE | |
| TSC2 | P49815 | S939 | SFRARSTSLNERPKS | |
| YBX1 | P67809 | S102 | NPRKYLRSVGDGETV |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
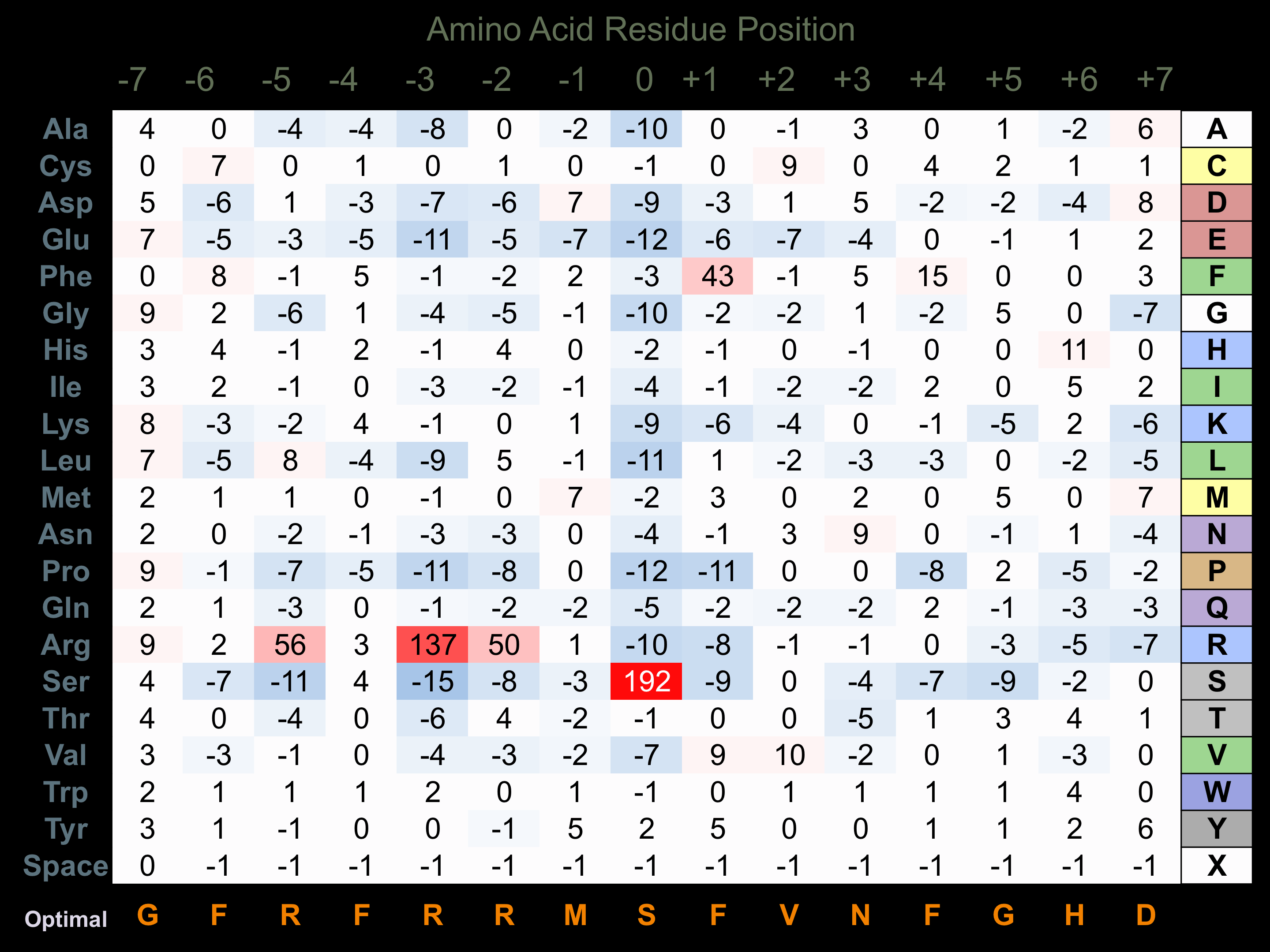
Matrix Type:
Experimentally derived from alignment of 63 known protein substrate phosphosites and 52 peptides phosphorylated by recombinant RSK1 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, neurological disorders
Specific Diseases (Non-cancerous):
Coffin-Lowry syndrome (CLS)
Specific Cancer Types:
Tuberous sclerosis (TSC)
Comments:
RSK1 may be a tumour requiring protein (TRP). The active form of the protein kinase normally acts to promote tumour cell proliferation. Abnormal activation of RSK1 expression has been linked to many human diseases, including breast and prostate cancers. Inhibition of RSK signalling enhances the effects of enzalutamide prostate cancer therapy.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +168, p<0.015); Cervical cancer (%CFC= -68, p<0.0001); Classical Hodgkin lymphomas (%CFC= +77, p<0.002); and Large B-cell lymphomas (%CFC= +86, p<0.0001). The COSMIC website notes an up-regulated expression score for RSK1 in diverse human cancers of 300, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 230 for this protein kinase in human cancers was 3.8-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 24903 diverse cancer specimens. This rate is only -32 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.41 % in 805 skin cancers tested; 0.31 % in 575 stomach cancers tested; 0.24 % in 1093 large intestine cancers tested.
Frequency of Mutated Sites:
None > 2 in 20,186 cancer specimens
Comments:
Only 4 deletions, no insertions or complex mutations.

