Nomenclature
Short Name:
JNK2
Full Name:
Mitogen-activated protein kinase 9
Alias:
- C-Jun N-terminal kinase 2
- EC 2.7.11.24
- MAPK9
- MK09
- P54a
- p54aSAPK; PRKM9; Stress-activated protein kinase JNK2; JNK2A, JNK2alpha; JNK2B, JNK2beta
- JNK2-alpha-2
- JNK-55
- Jun kinase
- Kinase JNK2
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
MAPK
SubFamily:
JNK
Specific Links
Structure
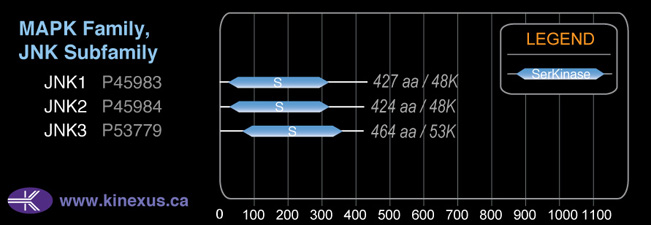
Mol. Mass (Da):
48139
# Amino Acids:
424
# mRNA Isoforms:
5
mRNA Isoforms:
48,311 Da (424 AA; P45984-4); 48,139 Da (424 AA; P45984); 44,223 Da (382 AA; P45984-3); 44,051 Da (382 AA; P45984-2); 27,334 Da (242 AA; P45984-5)
4D Structure:
Interacts with MECOM By similarity. Binds to at least four scaffolding proteins, MAPK8IP1/JIP-1, MAPK8IP2/JIP-2, MAPK8IP3/JIP-3/JSAP1 and SPAG9/MAPK8IP4/JIP-4. These proteins also bind other components of the JNK signaling pathway. Interacts with NFATC4.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 26 | 321 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K250.
Serine phosphorylated:
S129+, S155, S292, S311, S407.
Threonine phosphorylated:
T178+, T183+, T188-, T404.
Tyrosine phosphorylated:
Y185+, Y357.
Ubiquitinated:
K166, K250, K353.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 30
30
966
38
916
 3
3
84
18
49
 7
7
229
12
286
 24
24
766
134
1031
 28
28
901
35
665
 2
2
72
89
61
 10
10
334
45
523
 28
28
917
54
1468
 17
17
538
17
451
 4
4
144
125
147
 3
3
95
35
154
 21
21
684
206
668
 3
3
105
34
148
 2
2
49
15
42
 10
10
314
33
421
 3
3
112
21
55
 3
3
91
347
74
 7
7
220
22
460
 6
6
185
111
199
 21
21
681
137
624
 8
8
246
31
306
 5
5
145
31
157
 6
6
197
22
211
 5
5
163
23
270
 5
5
176
31
248
 24
24
784
86
833
 4
4
122
37
215
 4
4
132
23
193
 4
4
117
23
166
 5
5
146
42
146
 26
26
833
24
711
 100
100
3218
41
6867
 3
3
81
81
222
 33
33
1047
83
820
 3
3
89
48
89
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 42.9
42.9
57.8
100 84.7
84.7
91.5
94 -
-
-
99 -
-
-
99 99.3
99.3
100
97 -
-
-
- 96.7
96.7
97.4
99 98.8
98.8
98.8
99 -
-
-
- 97.9
97.9
99.1
- 88.9
88.9
90.1
99 79.8
79.8
87.8
- 71.7
71.7
81.8
86 -
-
-
- 65.1
65.1
76.2
- -
-
-
- 55.7
55.7
66.5
53 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | TP53 - P04637 |
| 2 | MAPK8IP3 - Q9UPT6 |
| 3 | SH3BP5 - O60239 |
| 4 | MAPK8IP1 - Q9UQF2 |
| 5 | TOB1 - P50616 |
| 6 | DUSP4 - Q13115 |
| 7 | ATF2 - P15336 |
| 8 | JUND - P17535 |
| 9 | NFATC3 - Q12968 |
| 10 | RB1 - P06400 |
| 11 | IRS1 - P35568 |
| 12 | MAP2K4 - P45985 |
| 13 | CDC42 - P60953 |
| 14 | ELK3 - P41970 |
| 15 | JDP2 - Q8WYK2 |
Regulation
Activation:
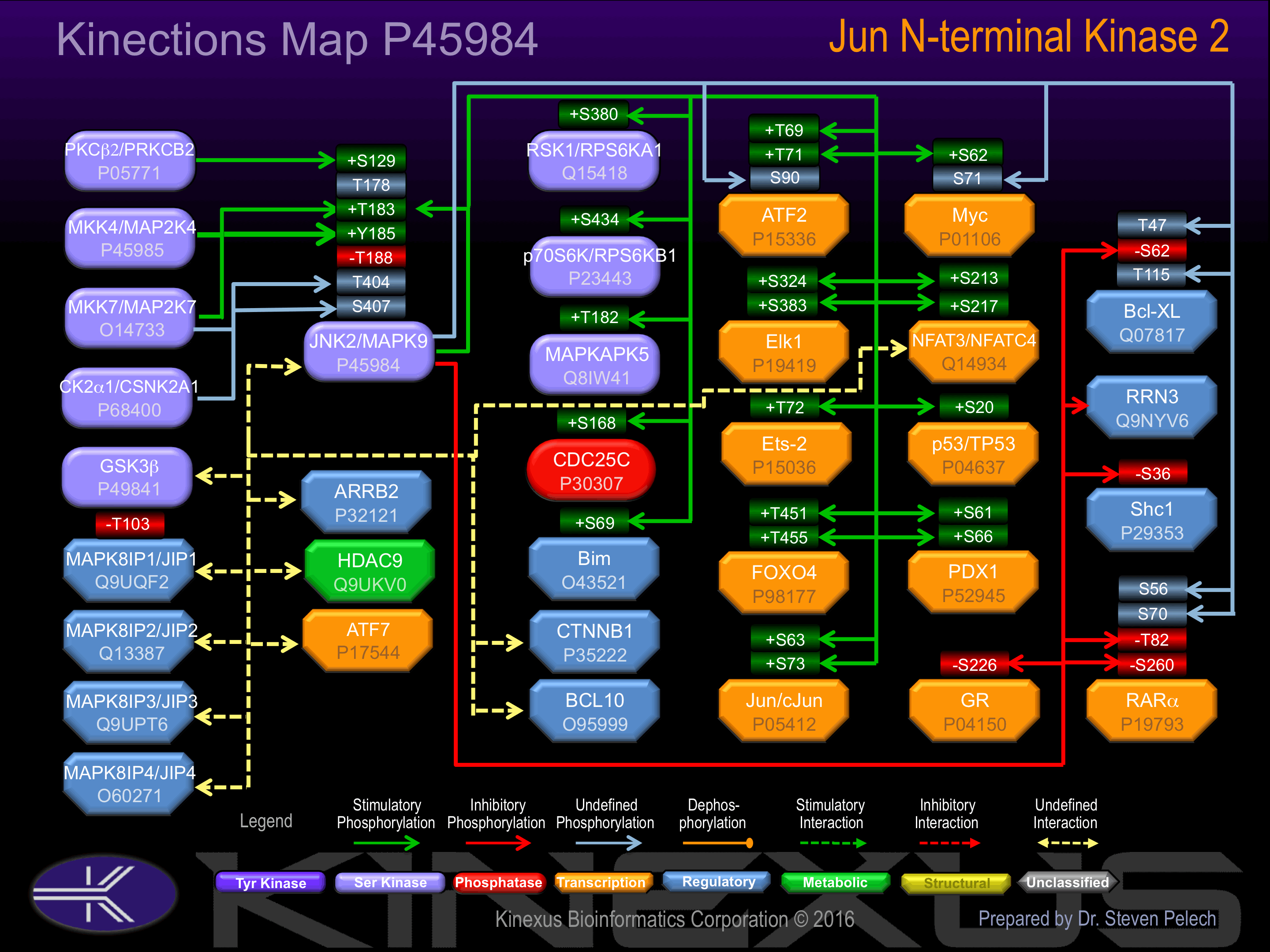
Phosphorylation of Thr-183 and Tyr-185 by either of two dual specificity kinases, MAP2K4 and MAP2K7, increases phosphotransferase activity. Phosphorylation of Ser-129 also increases phosphotransferase activity.
Inhibition:
Inhibited by dual specificity phosphatases, such as DUSP1, which target the Thr-183 and Tyr-185 activatory phosphorylation sites.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| PKCb | P05771 | S129 | ELDHERMSYLLYQML | + |
| MEK7 | O14733 | T183 | ACTNFMMTPYVVTRY | + |
| MEK4 | P45985 | T183 | ACTNFMMTPYVVTRY | + |
| JNK2 | P45984 | T183 | ACTNFMMTPYVVTRY | + |
| MEK7 | O14733 | Y185 | TNFMMTPYVVTRYYR | + |
| MEK4 | P45985 | Y185 | TNFMMTPYVVTRYYR | + |
| MEK7 | O14733 | T404 | SSMSTEQTLASDTDS | |
| CK2a1 | P68400 | T404 | SSMSTEQTLASDTDS | |
| MEK7 | O14733 | S407 | STEQTLASDTDSSLD | |
| CK2a1 | P68400 | S407 | STEQTLASDTDSSLD |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| APLP2 | Q06481 | T736 | VEVDPMLTPEERHLN | |
| APP | P05067 | T743 | VEVDAAVTPEERHLS | |
| ATF2 | P15336 | S90 | GLFNELASPFENEFK | |
| ATF2 | P15336 | T69 | SVIVADQTPTPTRFL | + |
| ATF2 | P15336 | T71 | IVADQTPTPTRFLKN | + |
| Bcl-xL | Q07817 | T115 | LTSQLHITPGTAYQS | |
| Bcl-xL | Q07817 | T47 | GTESEMETPSAINGN | |
| Bim | O43521 | S69 | GPLAPPASPGPFATR | |
| Cdc25C | P30307 | S168 | SEMKYLGSPITTVPK | + |
| CTNNB1 | P35222 | S191 | SRHAIMRSPQMVSAI | |
| CTNNB1 | P35222 | S605 | LFVQLLYSPIENIQR | |
| Doublecortin | O43602 | S415 | SPISTPTSPGSLRKH | |
| Doublecortin | O43602 | T402 | TSSSQLSTPKSKQSP | |
| Doublecortin | O43602 | T412 | SKQSPISTPTSPGSL | |
| Elk-1 | P19419 | S324 | RDLELPLSPSLLGGP | + |
| Elk-1 | P19419 | S383 | IHFWSTLSPIAPRSP | + |
| Ets-2 | P15036 | T72 | NCELPLLTPCSKAVM | + |
| FOXO4 | P98177 | T451 | PIPKALGTPVLTPPT | + |
| FOXO4 | P98177 | T455 | ALGTPVLTPPTEAAS | + |
| GR | P04150 | S226 | IDENCLLSPLAGEDD | - |
| H2AX | P16104 | S139 | GKKATQASQEY____ | |
| H3.2 | P84228 | S29 | ATKAARKSAPATGGV | |
| HSF1 | Q00613 | S363 | DTEGRPPSPPPTSTP | |
| JDP-2 | Q8WYK2 | T148 | VRTDSVKTPESEGNP | |
| JIP1 | Q9UQF2 | T103 | LIDATGDTPGAEDDE | - |
| JNK2 (MAPK9) | P45984 | T183 | ACTNFMMTPYVVTRY | + |
| Jun (c-Jun) | P05412 | S63 | KNSDLLTSPDVGLLK | + |
| Jun (c-Jun) | P05412 | S73 | VGLLKLASPELERLI | + |
| MAPKAPK5 | Q8IW41 | T182 | IDQGDLMTPQFTPYY | + |
| Myc | P01106 | S62 | LLPTPPLSPSRRSGL | |
| Myc | P01106 | S71 | SRRSGLCSPSYVAVT | |
| NFAT3 | Q14934 | S213 | ASRFGLGSPLPSPRA | + |
| NFAT3 | Q14934 | S217 | GLGSPLPSPRASPRP | + |
| NFAT4 | Q12968 | S163 | SYRESSLSPSPASSI | - |
| NFAT4 | Q12968 | S165 | RESSLSPSPASSISS | - |
| p53 | P04637 | S20 | PLSQETFSDLWKLLP | + |
| p70S6K (RPS6KB1) | P23443 | S434 | SFEPKIRSPRRFIGS | + |
| PDX1 | P52945 | S61 | LGALEQGSPPDISPY | + |
| PDX1 | P52945 | S66 | QGSPPDISPYEVPPL | + |
| PSEN1 | P49768 | S319 | NSKYNAESTERESQD | |
| PSEN1 | P49768 | T320 | SKYNAESTERESQDT | |
| RSK1 (RPS6KA1) | Q15418 | S380 | HQLFRGFSFVATGLM | + |
| RXRa | P19793 | S260 | NMGLNPSSPNDPVTN | - |
| RXRa | P19793 | S56 | SPISTLSSPINGMGP | |
| RXRa | P19793 | S70 | PPFSVISSPMGPHSM | |
| RXRa | P19793 | T82 | HSMSVPTTPTLGFST | - |
| SH3BP5 | O60239 | S351 | PGSLDLPSPVSLSEF | |
| Shc1 | P29353 | S36 | TPPEELPSPSASSLG | - |
| Tau iso8 | P10636-8 | S202 | SGYSSPGSPGTPGSR | |
| Tau iso8 | P10636-8 | T181 | KTPPAPKTPPSSGEP | |
| Tau iso8 | P10636-8 | T212 | TPGSRSRTPSLPTPP | |
| Tau iso8 | P10636-8 | T217 | SRTPSLPTPPTREPK | |
| Tau iso8 | P10636-8 | T231 | KKVAVVRTPPKSPSS | |
| TIF-IA | Q9NYV6 | T200 | IARYVPSTPWFLMPI | |
| TOB1 | P50616 | S152 | PASSVSSSPSPPFGH | |
| TOB1 | P50616 | S154 | SSVSSSPSPPFGHSA | |
| TOB1 | P50616 | S164 | FGHSAAVSPTFMPRS |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
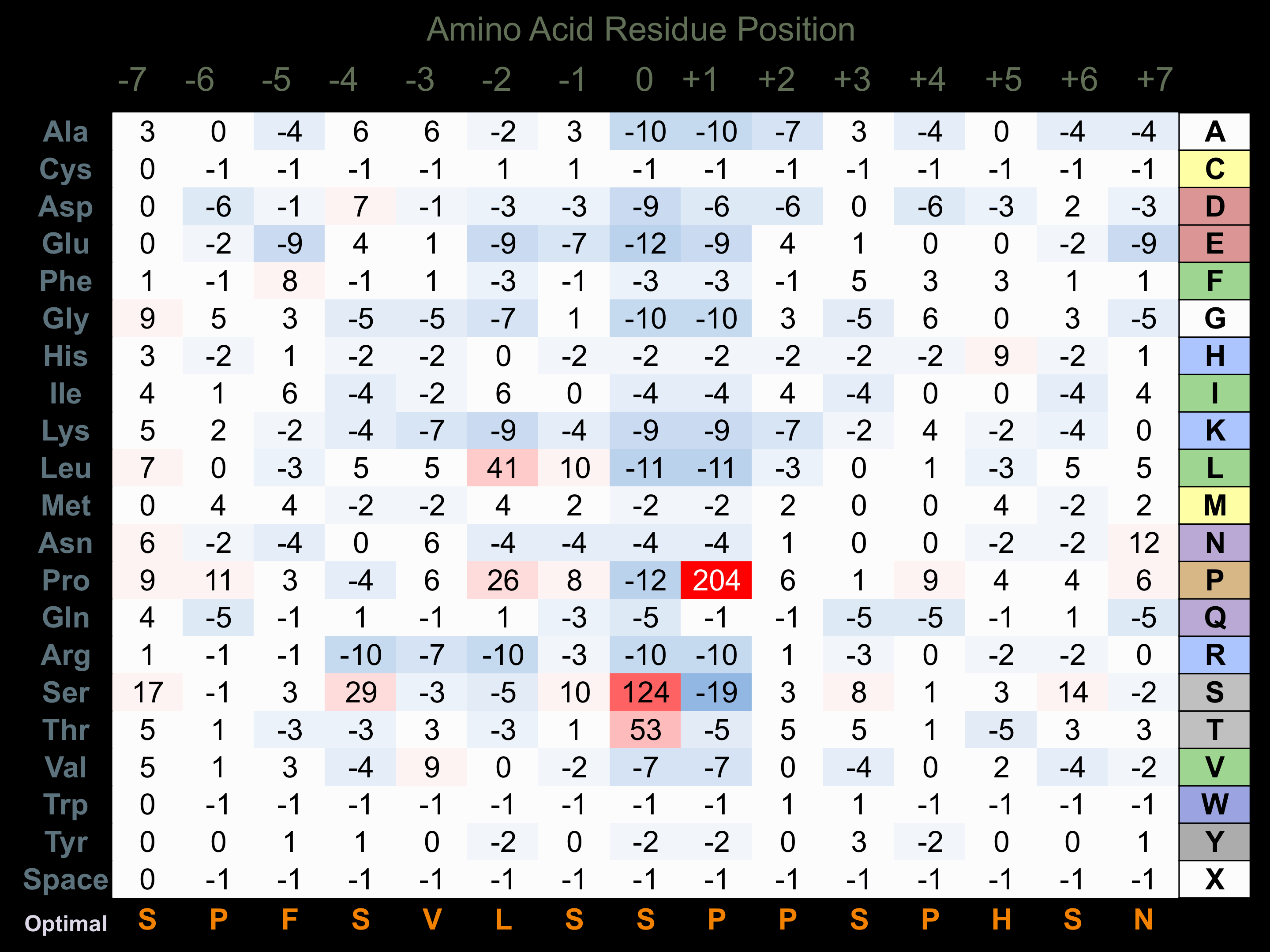
Matrix Type:
Experimentally derived from alignment of 64 known protein substrate phosphosites. Note that additional binding sites on JNK substrates with D motifs (consensus=R-P-t-s/r/t-L-p-L or K/r-x-s-L-s-L/i/v-s-l/p) facilitate higher selectivity for phosphorylation by this protein kinase.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Breast cancer
Comments:
JNK2 has been shown, in a cancer cell line to have a role in breast cancer cell motility.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in human Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +152, p<0.0001). The COSMIC website notes an up-regulated expression score for JNK2 in diverse human cancers of 563, which is 1.2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 230 for this protein kinase in human cancers was 3.8-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25492 diverse cancer specimens. This rate is very similar (+ 5% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.54 % in 1093 large intestine cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: G227C (4); G35E (3).
Comments:
Only 3 deletions, and no insertions or complex mutations are noted on the COSMIC website.