Nomenclature
Short Name:
MAPKAPK5
Full Name:
MAP kinase-activated protein kinase 5
Alias:
- EC 2.7.11.1
- MAP kinase-activated protein kinase 5
- MAPKAP kinase 5
- PRAK
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
MAPKAPK
SubFamily:
MAPKAPK
Specific Links
Structure
Mol. Mass (Da):
54,220
# Amino Acids:
473
# mRNA Isoforms:
2
mRNA Isoforms:
54,220 Da (473 AA; Q8IW41); 54,035 Da (471 AA; Q8IW41-2)
4D Structure:
Interacts with SQSTM1
1D Structure:
Subfamily Alignment
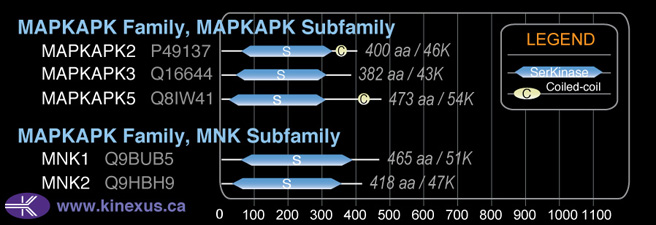
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 22 | 304 | Pkinase |
| 409 | 440 | Coiled-coil |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S115, S206, S212, S348, S354, S469, S472.
Threonine phosphorylated:
T182+, T186+, T214, T368.
Tyrosine phosphorylated:
Y188+, Y189+.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 17
17
849
16
1163
 1.2
1.2
57
10
22
 2
2
112
14
54
 9
9
450
59
881
 15
15
735
14
715
 1
1
50
37
14
 7
7
352
19
542
 15
15
746
33
1647
 15
15
763
10
709
 2
2
103
58
68
 1.4
1.4
71
26
39
 11
11
540
108
602
 1.5
1.5
73
25
51
 0.7
0.7
35
8
27
 2
2
104
24
76
 2
2
83
8
44
 1
1
48
112
26
 2
2
106
20
77
 2
2
109
56
91
 12
12
583
56
621
 2
2
96
23
69
 2
2
82
25
46
 2
2
106
15
68
 2
2
86
21
44
 2
2
121
23
90
 10
10
471
41
632
 2
2
96
28
65
 2
2
94
20
57
 2
2
93
21
61
 0.3
0.3
16
14
19
 18
18
911
18
591
 100
100
4950
21
7769
 27
27
1326
56
1374
 15
15
763
26
555
 2
2
91
22
57
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
100 70.7
70.7
70.7
100 -
-
-
98 -
-
-
97 58
58
58.3
98.5 -
-
-
- 97.2
97.2
98.7
97 34.4
34.4
52.8
96 -
-
-
- 57.2
57.2
58.1
- 21.1
21.1
35.3
96 34
34
52
91 87
87
94.2
88 -
-
-
- 32.3
32.3
49.2
- -
-
-
- 34.6
34.6
52.4
- 56
56
72.3
- -
-
-
- -
-
-
- -
-
-
- 21.1
21.1
37.3
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PLA2G4A - P47712 |
| 2 | TP53 - P04637 |
| 3 | MYH11 - P35749 |
| 4 | EIF4EBP1 - Q13541 |
| 5 | MAPK12 - P53778 |
| 6 | MAPK11 - Q15759 |
| 7 | HSPB1 - P04792 |
| 8 | MAPK1 - P28482 |
| 9 | HSPB2 - Q16082 |
| 10 | MAPK9 - P45984 |
| 11 | MAPK14 - Q16539 |
| 12 | GYS1 - P13807 |
| 13 | EIF4E - P06730 |
| 14 | ATF2 - P15336 |
| 15 | MBP - P02686 |
Regulation
Activation:
p38 alpha and beta-dependent phosphorylation increases its activity. Activated by stress-related extracellular stimuli; such as H2O2, arsenite, anisomycin TNF alpha and also PMA and the calcium ionophore A23187; but to a lesser extent. In vitro, activated by SQSTM1.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Akt1 (PKBa) | P31749 | S473 | RPHFPQFSYSASGTA | + |
| cPLA2 | P47712 | S727 | RQNPSRCSVSLSNVE | + |
| eIF4E | P07630 | S209 | DTATKSGSTTKNRFV | + |
| HSP27 (HSPB1) | P04792 | S15 | FSLLRGPSWDPFRDW | ? |
| HSP27 (HSPB1) | P04792 | S78 | PAYSRALSRQLSSGV | + |
| HSP27 (HSPB1) | P04792 | S82 | RALSRQLSSGVSEIR | ? |
| p53 (TP53) | P04637 | S37 | NVLSPLPSQAMDDLM | + |
| TH | P07101 | S19 | KGFRRAVSELDAKQA |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
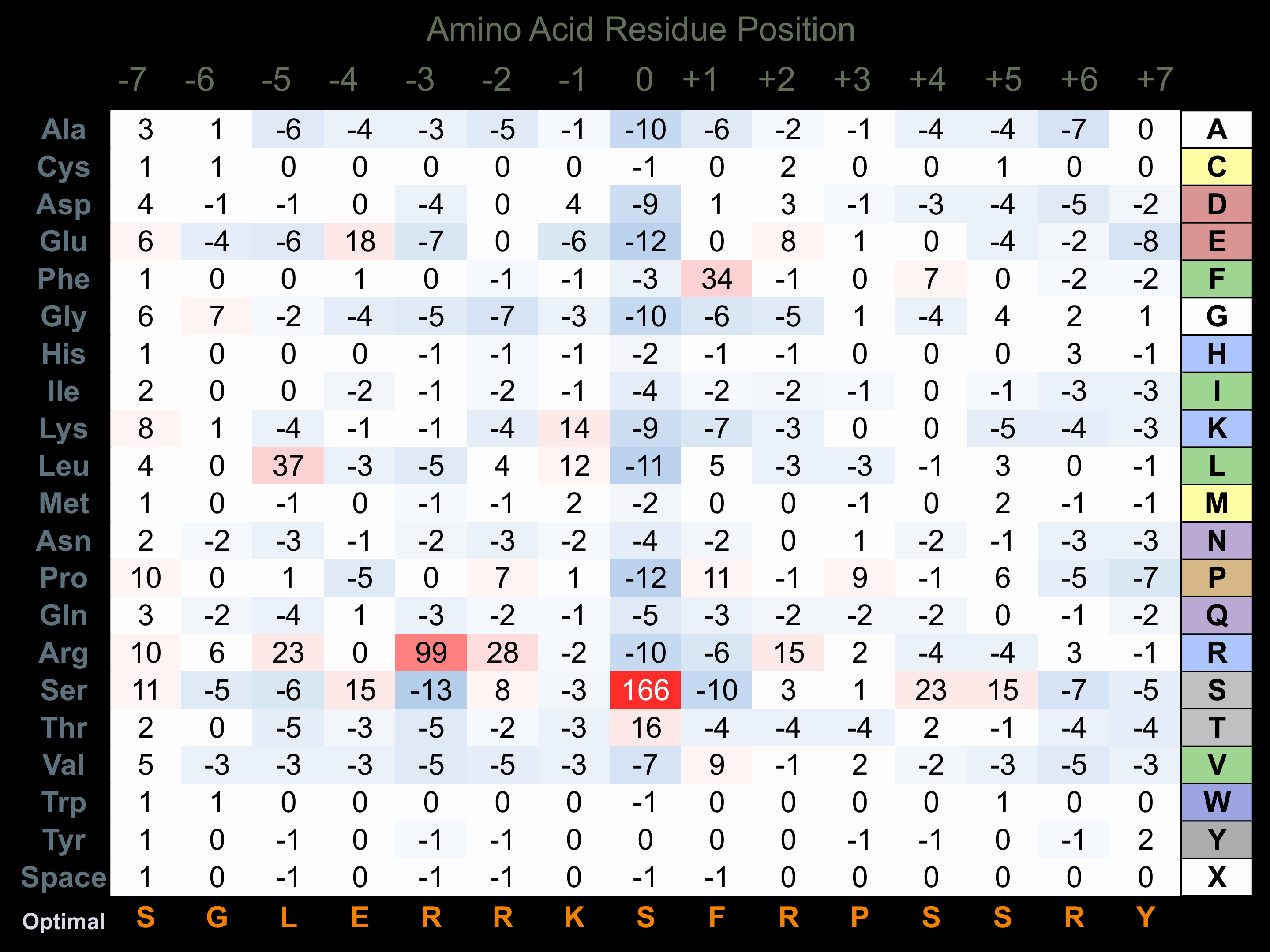
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Neurological disorders
Specific Diseases (Non-cancerous):
Alzheimer's disease (AD)
Comments:
The plasma level of MAPKAPK5 protein was found to positively associate with earlier detection of Alzheimer's Disease in a twin study.
Comments:
MAPKAPK5 is a tumour suppressor protein (TSP) that mediates Ras-induced senescence and phosphorylates p53/TP53.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +133, p<0.001); Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= +79, p<0.091); and Prostate cancer - primary (%CFC= +114, p<0.0007). The COSMIC website notes an up-regulated expression score for MAPKAPK5 in diverse human cancers of 766, which is 1.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 77 for this protein kinase in human cancers was 1.3-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. A K51M mutation abrogates MAPKAPK5's phosphotransferase activity. A T182A substitution leads to no p38-beta/MAPK11-induced activation. In addition, T182D, S212D and L337G mutations are associated with elevated phosphotransferase activity.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 25183 diverse cancer specimens. This rate is only -23 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.24 % in 1512 liver cancers tested; 0.18 % in 603 endometrium cancers tested; 0.18 % in 238 bone cancers tested; 0.15 % in 864 skin cancers tested; 0.12 % in 710 oesophagus cancers tested; 0.08 % in 555 stomach cancers tested; 0.08 % in 273 cervix cancers tested; 0.07 % in 942 upper aerodigestive tract cancers tested; 0.05 % in 939 prostate cancers tested.
Frequency of Mutated Sites:
None > 2 in 20,466 cancer specimens
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.

