Nomenclature
Short Name:
CDK2
Full Name:
Cyclin-dependent kinase 2
Alias:
- Cyclin-dependent kinase 2
- EC 2.7.11.22
- Kinase Cdk2
- P33 protein kinase
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
CDK
SubFamily:
CDC2
Specific Links
Structure
Mol. Mass (Da):
33,930
# Amino Acids:
298
# mRNA Isoforms:
2
mRNA Isoforms:
33,930 Da (298 AA; P24941); 30,035 Da (264 AA; P24941-2)
4D Structure:
Found in a complex with CABLES1, CCNA1 and CCNE1. Interacts with CABLES1. Interacts with UHRF2. Part of a complex consisting of UHRF2, CDK2 and CCNE1. Interacts with the Speedy/Ringo proteins SPDYA and SPDYC. Found in a complex with both SPDYA and CDKN1B/KIP1.
3D Structure:
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 4 | 286 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K6.
Serine phosphorylated:
S46.
Sumoylated:
K33.
Threonine phosphorylated:
T14-, T39, T137, T158+, T160+, T165-.
Tyrosine phosphorylated:
Y15-, Y19+, Y159+, Y168-, Y179+, Y269.
Ubiquitinated:
K9, K20, K24, K33, K56, K129, K142, K237, K250, K273, K278, K291.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 60
60
802
42
1010
 4
4
57
20
52
 7
7
90
8
65
 59
59
794
122
3052
 49
49
663
36
602
 24
24
322
119
1181
 21
21
278
47
575
 100
100
1344
48
3005
 25
25
336
24
263
 8
8
102
113
138
 6
6
77
35
92
 50
50
668
236
605
 4
4
59
41
60
 5
5
71
18
86
 8
8
104
26
115
 3
3
38
22
35
 11
11
146
207
130
 5
5
67
19
62
 4
4
59
125
69
 32
32
436
162
498
 6
6
80
23
85
 8
8
113
30
154
 6
6
85
19
54
 6
6
87
19
72
 17
17
234
23
293
 83
83
1118
71
2448
 6
6
77
44
75
 7
7
98
19
86
 7
7
92
20
83
 8
8
101
42
103
 43
43
575
30
657
 38
38
510
51
498
 88
88
1185
91
1875
 48
48
651
83
586
 4
4
55
57
61
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 69.8
69.8
79.7
100 99.7
99.7
99.7
100 -
-
-
99 -
-
-
98 99
99
99.7
99 -
-
-
- 85.3
85.3
85.8
99 98.7
98.7
99.7
99 -
-
-
- -
-
-
- 65.3
65.3
77.9
93 88.9
88.9
95
90 90.3
90.3
95
90 -
-
-
- 62.7
62.7
74.8
66 69.9
69.9
82.3
- 56.9
56.9
69.9
- 75.9
75.9
88.6
- 66.4
66.4
83.2
- 66.8
66.8
82.2
- -
-
-
66 67.1
67.1
81.5
69 62.4
62.4
79.2
65 -
-
-
65
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CCNA2 - P20248 |
| 2 | CCNA1 - P78396 |
| 3 | CDKN3 - Q16667 |
| 4 | CCNE1 - P24864 |
| 5 | ORC2L - Q13416 |
| 6 | CCNE2 - O96020 |
| 7 | MCM3 - P25205 |
| 8 | CDKN1B - P46527 |
| 9 | CCNB2 - O95067 |
| 10 | CCNB3 - Q8WWL7 |
| 11 | CKS1B - P61024 |
| 12 | SKP2 - Q13309 |
| 13 | BRCA1 - P38398 |
| 14 | RB1 - P06400 |
| 15 | RBL1 - P28749 |
Regulation
Activation:
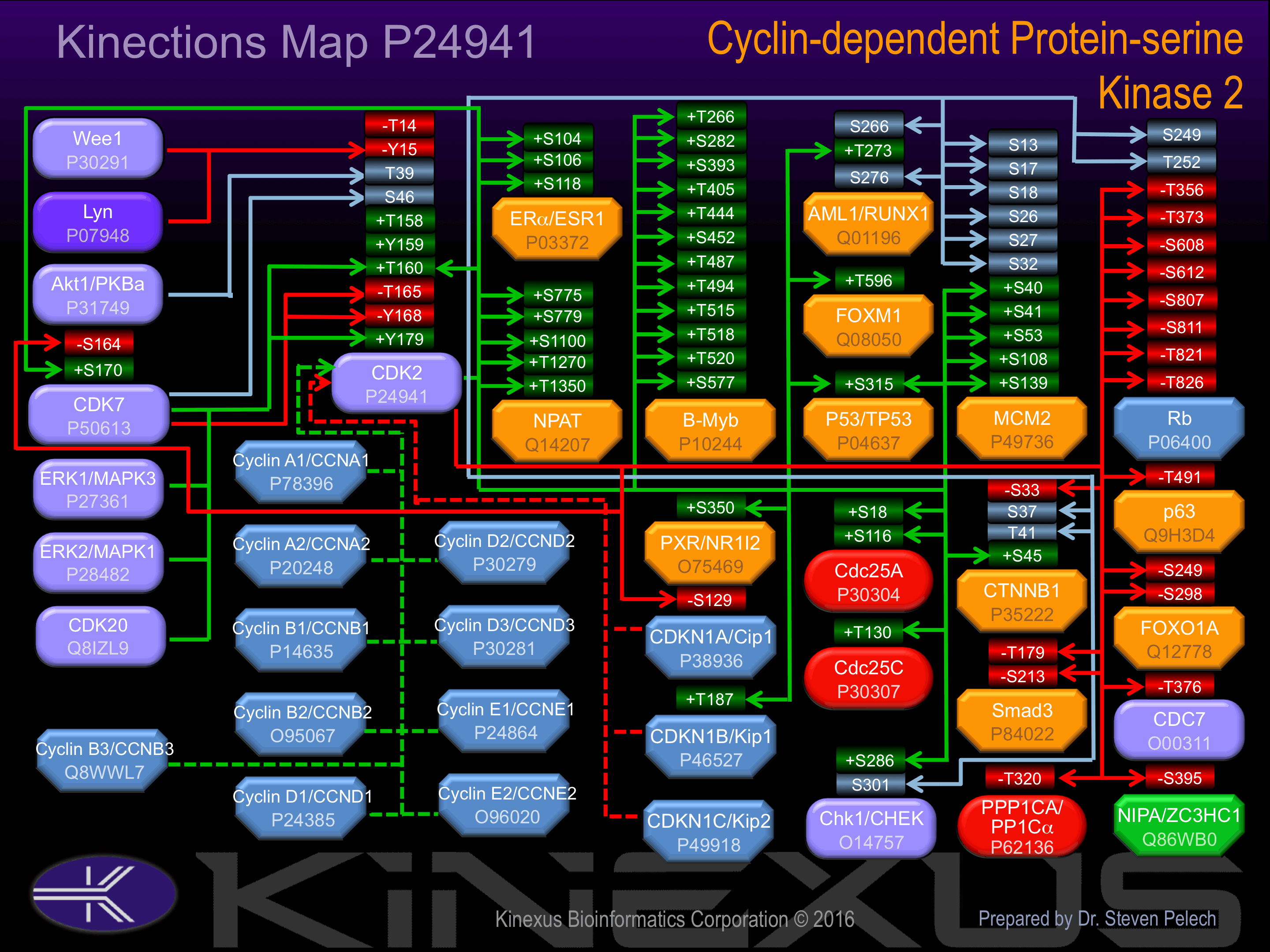
Phosphorylation at Thr-160 increases phosphotransferase activity. Phosphorylation at Tyr-179 increases phosphotransferase activity and interaction with CCNA2. Cyclin A2 stimulates the catalytic activity of CDK2.
Inhibition:
Phosphorylation at Thr-14 and Tyr-15 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| LYN | P07948 | Y15 | EKIGEGTYGVVYKAR | - |
| WEE1 | P30291 | Y15 | EKIGEGTYGVVYKAR | - |
| Akt1 | P31749 | T39 | LKKIRLDTETEGVPS | ? |
| CDK7 | P50613 | S46 | TETEGVPSTAIREIS | |
| CDK7 | P50613 | T160 | GVPVRTYTHEVVTLW | + |
| ERK1 | P27361 | T160 | GVPVRTYTHEVVTLW | + |
| CDK2 | P24941 | T160 | GVPVRTYTHEVVTLW | + |
| ERK2 | P28482 | T160 | GVPVRTYTHEVVTLW | + |
| CCRK | Q8IZL9 | T160 | GVPVRTYTHEVVTLW | + |
| CDK7 | P50613 | T165 | TYTHEVVTLWYRAPE | - |
| CDK7 | P50613 | Y168 | HEVVTLWYRAPEILL | - |
| CDK7 | P50613 | Y179 | EILLGCKYYSTAVDI | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| AML1 (RUNX1) | Q01196 | S266 | QYLGSIASPSVHPAT | |
| AML1 (RUNX1) | Q01196 | S276 | VHPATPISPGRASGM | |
| AML1 (RUNX1) | Q01196 | T273 | SPSVHPATPISPGRA | + |
| ANKRD17 | O75179 | S2042 | KEHYPVSSPSSPSPP | |
| ANKRD17 | O75179 | S2045 | YPVSSPSSPSPPAQP | |
| ANKRD17 | O75179 | S2401 | SSGVRAPSPAPSSVP | |
| ATRIP | Q8WXE1 | S224 | APSVSHVSPRKNPSV | |
| Axin | O15169 | S486 | LRTPGRQSPGPGHRS | |
| Axin | O15169 | S493 | SPGPGHRSPDSGHVA | |
| Axin | O15169 | T481 | HVQRVLRTPGRQSPG | |
| B-Myb (MYBL2) | P10244 | S282 | KREDQEGSPPETSLP | + |
| B-Myb (MYBL2) | P10244 | S393 | RGELIPISPSTEVGG | + |
| B-Myb (MYBL2) | P10244 | S452 | PVKTLPFSPSQFLNF | + |
| B-Myb (MYBL2) | P10244 | S577 | RKPGLRRSPIKKVRK | + |
| B-Myb (MYBL2) | P10244 | T266 | TDLDAVRTPEPLEEF | + |
| B-Myb (MYBL2) | P10244 | T405 | VGGSGIGTPPSVLKR | + |
| B-Myb (MYBL2) | P10244 | T444 | NSLTPKSTPVKTLPF | + |
| B-Myb (MYBL2) | P10244 | T487 | SQKVVVTTPLHRDKT | + |
| B-Myb (MYBL2) | P10244 | T494 | TPLHRDKTPLHQKHA | + |
| B-Myb (MYBL2) | P10244 | T515 | QKYSMDNTPHTPTPF | + |
| B-Myb (MYBL2) | P10244 | T518 | SMDNTPHTPTPFKNA | + |
| B-Myb (MYBL2) | P10244 | T520 | DNTPHTPTPFKNALE | + |
| BARD1 | Q99728 | S148 | NSIKMWFSPRSKKVR | |
| BARD1 | Q99728 | S251 | SQPSVISSPQINGEI | |
| BARD1 | Q99728 | S288 | PLAEQIESPDTKSRN | |
| BARD1 | Q99728 | T299 | KSRNEVVTPEKVCKN | |
| BRCA1 | P38398 | S1497 | EPGVERSSPSKCPSL | |
| BTG1 | P62324 | S159 | ELLLGRTSPSKNYNM | |
| C-EBPb | P17676 | T235 | SSSSPPGTPSPADAK | |
| Cdc25A | P30304 | S116 | PQKLLGCSPALKRSH | + |
| Cdc25A | P30304 | S18 | RRLLFACSPPPASQP | + |
| Cdc25C | P30307 | T130 | PAQLLCSTPNGLDRG | + |
| Cdc6 | Q99741 | S106 | DNQLTIKSPSKRELA | |
| Cdc6 | Q99741 | S54 | RVKALPLSPRKRLGD | |
| Cdc6 | Q99741 | S74 | TPHLPPCSPPKQGKK | |
| CDC7 | O00311 | T376 | QVAPRAGTPGFRAPE | - |
| CDK2 | P24941 | T160 | GVPVRTYTHEVVTLW | + |
| CDK7 | P50613 | S164 | GLAKSFGSPNRAYTH | - |
| CDK7 | P50613 | T170 | GSPNRAYTHQVVTRW | + |
| Cdt1 | Q9H211 | S31 | KLACRTPSPARPALR | |
| Cdt1 | Q9H211 | T29 | PPKLACRTPSPARPA | |
| CDX2 | Q99626 | S283 | RSVPEPLSPVSSLQA | |
| Chk1 (CHEK1) | O14757 | S286 | TSGGVSESPSGFSKH | + |
| Chk1 (CHEK1) | O14757 | S301 | IQSNLDFSPVNSASS | ? |
| Cip1 (p21, CDKN1A) | P38936 | S129 | SGEQAEGSPGGPGDS | - |
| Coilin | P38432 | S184 | SVCSKRKSPKKKEKC | |
| Ctip | Q99708 | T847 | FRYIPPNTPENFWEV | |
| CTNNB1 | P35222 | S33 | QQQSYLDSGIHSGAT | - |
| CTNNB1 | P35222 | S37 | YLDSGIHSGATTTAP | ? |
| CTNNB1 | P35222 | S45 | GATTTAPSLSGKGNP | + |
| CTNNB1 | P35222 | T41 | GIHSGATTTAPSLSG | ? |
| CTTN (Cortactin) | Q14247 | S405 | KTQTPPVSPAPQPTE | - |
| Cyclin A2 | P20248 | S154 | PMDGSFESPHTMDMS | |
| Cyclin E1 (CCNE1) | P24864 | S387 | LSEQNRASPLPSGLL | |
| Cyclin E1 (CCNE1) | P24864 | S399 | GLLTPPQSGKKQSSG | |
| Cyclin E1 (CCNE1) | P24864 | T395 | PLPSGLLTPPQSGKK | |
| DDX3 | O00571 | T203 | LTRYTRPTPVQKHAI | |
| DLG1 (SAP97) | Q12959 | S158 | FVSHSHISPIKPTEA | |
| DLG1 (SAP97) | Q12959 | S443 | FLGQTPASPARYSPV | |
| E2A | P15923 | S139 | LNSPGPLSPSGMKGT | |
| E2A | P15923 | S245 | PMLGGGSSPLPLPPG | |
| E2F5 | Q15329 | T261 | SQSLTPVTPQKSSMA | |
| eEF2 | P13639 | T435 | RIMGPNYTPGKKEDL | |
| ERa (ESR1) | P03372 | S104 | FPPLNSVSPSPLMLL | + |
| ERa (ESR1) | P03372 | S106 | PLNSVSPSPLMLLHP | + |
| ERa (ESR1) | P03372 | S118 | LHPPPQLSPFLQPHG | + |
| FOXM1 | Q08050 | T611 | ETLPISSTPSKSVLP | + |
| FOXO1A | Q12778 | S249 | EGGKSGKSPRRRAAS | - |
| FOXO1A | Q12778 | S298 | QFSKWPASPGSHSND | - |
| GANP | O60318 | S508 | FWHRKKISPNKKPFS | |
| GR | P04150 | S203 | DLEFSSGSPGKETNE | |
| GR | P04150 | S211 | PGKETNESPWRSDLL | ? |
| GRASP65 | Q9BQQ3 | S273 | DPLPGPGSPSHSAPD | |
| HIRA | P54198 | T555 | LSPSVLTTPSKIEPM | |
| hnRNP K | P61978 | S216 | ILDLISESPIKGRAQ | + |
| HR6A | P49459 | S120 | LDEPNPNSPANSQAA | |
| Id2 | Q02363 | S5 | ___MKAFSPVRSVRK | |
| Id3 | Q02535 | S5 | ___MKALSPVRGCYE | |
| IREB2 | P48200 | S157 | LQKAGKLSPLKVQPK | |
| LIG1 | P18858 | S51 | GVVSESDSPVKRPGR | |
| LIG1 | P18858 | S76 | EEEDEALSPAKGQKP | |
| LIG1 | P18858 | S91 | ALDCSQVSPPRPATS | |
| LIG3 | P49916 | S210 | TTTGQVTSPVKGASF | |
| LIG3 | P49916 | S913 | EKLATKSSPVKVGEK | |
| LIG3 | P49916 | T191 | LSSKAAGTPKKKAVV | |
| MAP2 | P11137 | S136 | ETANLPPSPPPSPAS | |
| MARCKS | P29966 | S26 | GEAAVASSPSKANGQ | |
| MARCKS | P29966 | S27 | GEAAVASSPSKANGQ | |
| MARCKS | P29966 | T150 | TPSPSNETPKKKKKR | |
| MCL1 | Q07820 | S64 | IGGSAGASPPSTLTP | |
| MCM2 | P49736 | S108 | DVEELTASQREAAER | + |
| MCM2 | P49736 | S13 | ESFTMASSPAQRRRG | ? |
| MCM2 | P49736 | S139 | RRGLLYDSDEEDEER | + |
| MCM2 | P49736 | S17 | RGNDPLTSSPGRSSR | |
| MCM2 | P49736 | S18 | GNDPLTSSPGRSSRR | |
| MCM2 | P49736 | S26 | RGNDPLTSSPGRSSR | |
| MCM2 | P49736 | S27 | GNDPLTSSPGRSSRR | ? |
| MCM2 | P49736 | S32 | RTDALTSSPGRDLPP | |
| MCM2 | P49736 | S40 | RRTDALTSSPGRDLP | + |
| MCM2 | P49736 | S41 | RTDALTSSPGRDLPP | + |
| MCM2 | P49736 | S53 | LPPFEDESEGLLGTE | + |
| MCM3 | P25205 | S611 | SSDTARTSPVTARTL | |
| MCM4 | P33991 | S3 | _____MSSPASTPSR | |
| MCM4 | P33991 | S32 | RSEDARSSPSQRRRG | |
| MCM4 | P33991 | S54 | ELQPMPTSPGVDLQS | |
| MCM4 | P33991 | T110 | PRSGVRGTPVRQRPD | |
| MCM4 | P33991 | T19 | GSRRGRATPAQTPRS | |
| MDM2 | Q00987 | T218 | SSSESTGTPSNPDLD | |
| MELF4 (MEF) | Q99607 | S648 | SPTPAPFSPFNPTSL | - |
| MELF4 (MEF) | Q99607 | T643 | SLLTRSPTPAPFSPF | |
| MyoD | P15172 | S200 | SGDSDASSPRSNCSD | |
| NF-YA | P23511 | S320 | GEGGRFFSPKEKDSP | |
| NF-YA | P23511 | S326 | FSPKEKDSPHMQDPN | |
| Nogo | Q9NQC3 | S15 | PLVSSSDSPPRPQPA | |
| NPAT | Q14207 | S1100 | VSFPNLDSPNVSSTL | + |
| NPAT | Q14207 | S775 | LPTIILSSPTKSPTK | + |
| NPAT | Q14207 | S779 | ILSSPTKSPTKNAEL | + |
| NPAT | Q14207 | T1270 | SDLPVPRTPGSGAGE | + |
| NPAT | Q14207 | T1350 | ISRTTSATPLKDNTQ | + |
| NPM1 | P06748 | S70 | EAMNYEGSPIKVTLA | |
| NPM1 | P06748 | T199 | VKKSIRDTPAKNAQK | |
| NPM1 | P06748 | T234 | SFKKQEKTPKTPKGP | |
| NRIF3 | Q13352 | S33 | KKSVITYSPTTGTCQ | |
| NRIF3 | Q13352 | S46 | CQMSLFASPTSSEEQ | |
| NSBP1 | P82970 | T31 | SAMLVPVTPEVKPKR | |
| p27Kip1 | P46527 | T187 | NAGSVEQTPKKPGLR | + |
| p53 | P04637 | S315 | LPNNTSSSPQPKKKP | + |
| p63 | Q9H3D4 | T491 | PQQRNALTPTTIPDG | - |
| p73 | O15350 | T86 | AASASPYTPEHAASV | |
| POLL | Q9UGP5 | S167 | ALLQTALSPPPPPTR | |
| POLL | Q9UGP5 | S177 | PPPTRPVSPPQKAKE | |
| POLL | Q9UGP5 | S230 | LEGDCEPSPAPAVLD | |
| POLL | Q9UGP5 | T553 | GPGRVLPTPTEKDVF | |
| PPP1CA | P62138 | T320 | NPGGRPITPPRNSAK | - |
| PR | P06401 | S162 | PATQRVLSPLMSRSG | ? |
| PR | P06401 | S190 | KVLPRGLSPARQLLL | ? |
| PR | P06401 | S213 | SGAPVKPSPQAAAVE | |
| PR | P06401 | S400 | GAEASARSPRSYLVA | ? |
| PR | P06401 | S676 | LSQRFTFSPGQDIQL | ? |
| PRKAR1A | P10644 | S83 | DSREDEISPPPPNPV | - |
| PTPN2 | P17706 | S304 | LSPAFDHSPNKIMTE | |
| PTPN6 (SHP1) | P29350 | S591 | DKEKSKGSLKRK___ | |
| PXR (NR1I2) | O75469 | S350 | MQAISLFSPDRPGVL | + |
| Rad9 | Q99638 | S328 | VLPSISLSPGPQPPK | ? |
| RanGAP1 | P46060 | S442 | STFLAFPSPEKLLRL | |
| RanGAP1 | P46060 | T409 | GQGEKSATPSRKILD | |
| Rb | P06400 | S249 | AVIPINGSPRTPRRG | |
| Rb | P06400 | S608 | TAADMYLSPVRSPKK | - |
| Rb | P06400 | S612 | MYLSPVRSPKKKGST | - |
| Rb | P06400 | S807 | PGGNIYISPLKSPYK | - |
| Rb | P06400 | S811 | IYISPLKSPYKISEG | - |
| Rb | P06400 | T252 | PINGSPRTPRRGQNR | |
| Rb | P06400 | T356 | DSFETQRTPRKSNLD | - |
| Rb | P06400 | T373 | VNVIPPHTPVRTVMN | - |
| Rb | P06400 | T821 | KISEGLPTPTKMTPR | - |
| Rb | P06400 | T826 | LPTPTKMTPRSRILV | - |
| Rb-like 2 | Q08999 | S1044 | YPFVRTGSPRRIQLS | |
| Rb-like 2 | Q08999 | S1068 | HKNETMLSPREKIFY | |
| Rb-like 2 | Q08999 | S1080 | IFYYFSNSPSKRLRE | |
| Rb-like 2 | Q08999 | S1112 | LLEDGSESPAKRICP | |
| Rb-like 2 | Q08999 | S413 | VRYIKENSPCVTPVS | |
| Rb-like 2 | Q08999 | S639 | DEICIAGSPLTPRRV | |
| Rb-like 2 | Q08999 | S662 | GLGRSITSPTTLYDR | |
| Rb-like 2 | Q08999 | S688 | RLFVENDSPSDGGTP | |
| Rb-like 2 | Q08999 | S952 | DSRSHQNSPTELNKD | |
| Rb-like 2 | Q08999 | T1097 | SMIRTGETPTKKRGI | |
| Rb-like 2 | Q08999 | T417 | KENSPCVTPVSTATH | |
| Rb-like 2 | Q08999 | T642 | CIAGSPLTPRRVTEV | |
| Rb-like 2 | Q08999 | T694 | DSPSDGGTPGRMPPQ | |
| RPL12 | P30050 | S38 | KIGPLGLSPKKVGDD | |
| SF3B1 | O75533 | T244 | GRAKGSETPGATPGS | |
| SF3B1 | O75533 | T248 | GSETPGATPGSKIWD | |
| SF3B1 | O75533 | T313 | HGSGWAETPRTDRGG | |
| SIRT2 | Q8IXJ6 | S368 | PNPSTSASPKKSPPP | |
| SKP2 | Q13309 | S64 | SNLGHPESPPRKRLK | |
| Smad3 | P84022 | S213 | NLSPNPMSPAHNNLD | - |
| Smad3 | P84022 | T179 | PQSNIPETPPPGYLS | - |
| Smad3 | P84022 | T8 | MSSILPFTPPIVKRL | |
| SNRNP70 | P08621 | S226 | YDERPGPSPLPHRDR | |
| TIF-IA | Q9NYV6 | S44 | LENDFFNSPPRKTVR | |
| TK1 | P04183 | S13 | LPTVLPGSPSKTRGQ | |
| UBF | P17480 | S389 | INKKQATSPASKKPA | |
| UBF | P17480 | S484 | ERGKLPESPKRAEEI | |
| ZBTB16 | Q05516 | S197 | SFGLSAMSPTKAAVD | |
| ZBTB16 | Q05516 | T282 | RGKEGPGTPTRSSVI | |
| ZC3HC1 (NIPA) | Q86WB0 | S395 | PGLEVPSSPLRKAKR | - |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
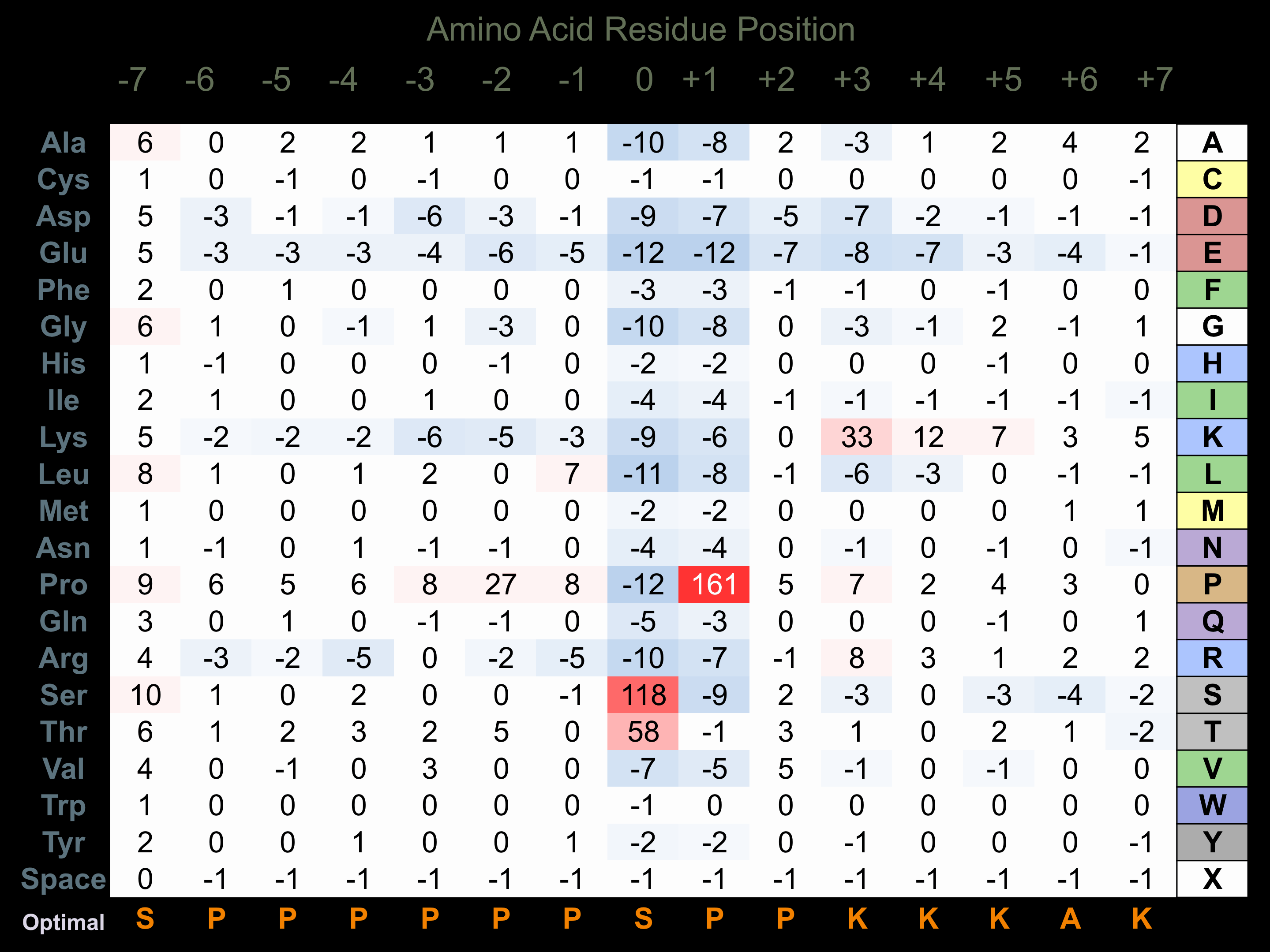
Matrix Type:
Experimentally derived from alignment of 543 known protein substrate phosphosites and 35 peptides phosphorylated by recombinant CDK2 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, skin and mucosal lesions
Specific Diseases (Non-cancerous):
Pemphigus vulgaris
Comments:
In pemphigus vulgaris skin and mucosal membrane lesions will develop, and CDK2 plays an essential role.
Specific Cancer Types:
Tetraploidy; Male germ cell tumours
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Colon mucosal cell adenomas (%CFC= +78, p<0.0001); Skin melanomas - malignant (%CFC= +512, p<0.0001); and Vulvar intraepithelial neoplasia (%CFC= +130, p<0.004). The COSMIC website notes an up-regulated expression score for CDK2 in diverse human cancers of 560, which is 1.2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 42 for this protein kinase in human cancers was 0.7-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. Mutations resulting in reduced CAK phosphorylation at the T160 activating phosphosite include K9F, and K88E + K89V, while a L166R mutation can result in reduced CAK phosphorylation and lower protein-serine/threonine phosphotransferase activity. Phosphotransferase activity can be fully removed with a T160A mutation. Phosphotransferase activity can be increased 2-fold with either a T14A, or a Y15F mutation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 25535 diverse cancer specimens. This rate is only -21 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.42 % in 1128 large intestine cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: P45L (3).
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.