Nomenclature
Short Name:
IKKa
Full Name:
Inhibitor of nuclear factor kappa-B kinase alpha subunit
Alias:
- CHUK
- I-kappa-B kinase 1
- IkBKA
- IKK1
- IKK-alpha
- NFKBIKA
- Conserved helix-loop-helix ubiquitous kinase
- EC 2.7.11.10
- I kappa-B kinase alpha
- IkappaB kinase
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
IKK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
84,654
# Amino Acids:
745
# mRNA Isoforms:
1
mRNA Isoforms:
84,640 Da (745 AA; O15111)
4D Structure:
Component of the I-kappa-B-kinase (IKK) core complex consisting of CHUK, IKBKB and IKBKG; probably four alpha/CHUK-beta/IKBKB dimers are associated with four gamma/IKBKG subunits. The IKK core complex seems to associate with regulatory or adapter proteins to form a IKK-signalosome holo-complex. Part of a complex composed of NCOA2, NCOA3, CHUK/IKKA, IKBKB, IKBKG and CREBBP. Part of a 70-90 kDa complex at least consisting of CHUK/IKKA, IKBKB, NFKBIA, RELA, IKBKAP and MAP3K14. Directly interacts with IKK-gamma/NEMO and TRPC4AP By similarity. May interact with TRAF2. Interacts with NALP2. May interact with MAVS/IPS1. Interacts with ARRB1 and ARRB2.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 15 | 313 | Pkinase |
| 455 | 476 | Leucine-zipper |
| 706 | 743 | IKKbetaNEMObind |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
S119, T179, K322, K438, T722.
Serine phosphorylated:
S119, S176+, S180+, S473.
Threonine phosphorylated:
T23+, T463+.
Tyrosine phosphorylated:
Y168, Y187+,Y500.
Ubiquitinated:
K296, K415.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 10
10
594
22
948
 1.4
1.4
85
10
120
 2
2
114
21
102
 4
4
243
66
265
 12
12
698
14
682
 0.6
0.6
34
37
19
 7
7
412
19
717
 13
13
784
41
1747
 7
7
435
10
438
 3
3
157
45
180
 2
2
131
34
142
 8
8
491
126
497
 2
2
110
32
114
 1.4
1.4
84
9
102
 2
2
139
31
120
 2
2
115
8
101
 3
3
189
114
95
 2
2
117
28
107
 2
2
142
63
157
 10
10
591
56
643
 3
3
206
30
187
 3
3
154
32
172
 3
3
158
22
147
 2
2
121
28
106
 3
3
193
30
231
 7
7
406
48
485
 2
2
104
35
116
 2
2
106
28
98
 2
2
138
28
123
 1.1
1.1
66
14
41
 18
18
1065
18
714
 100
100
5929
19
11003
 4
4
247
56
441
 20
20
1212
26
887
 2
2
139
22
134
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.7
99.7
99.9
100 99.6
99.6
99.9
99.5 -
-
-
98 -
-
-
- 97.7
97.7
98.8
98 -
-
-
- 95.6
95.6
97.5
95 49.1
49.1
66.7
95 -
-
-
- 80.1
80.1
87
- 80.1
80.1
88.9
81 72.6
72.6
85.6
74 62.9
62.9
77.8
64 -
-
-
- 24.4
24.4
45
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | NFKBIA - P25963 |
| 2 | RELA - Q04206 |
| 3 | NFKBIB - Q15653 |
| 4 | NFKB1 - P19838 |
| 5 | AKT1 - P31749 |
| 6 | CDC37 - Q16543 |
| 7 | TP53 - P04637 |
| 8 | MAP3K11 - Q16584 |
| 9 | SMAD2 - Q15796 |
| 10 | PRKDC - P78527 |
| 11 | ESR1 - P03372 |
| 12 | AKT2 - P31751 |
| 13 | BCL10 - O95999 |
| 14 | UBE2E3 - Q969T4 |
| 15 | TNFAIP3 - P21580 |
Regulation
Activation:
Activated by phosphorylation.at Thr-23 and Ser-176.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
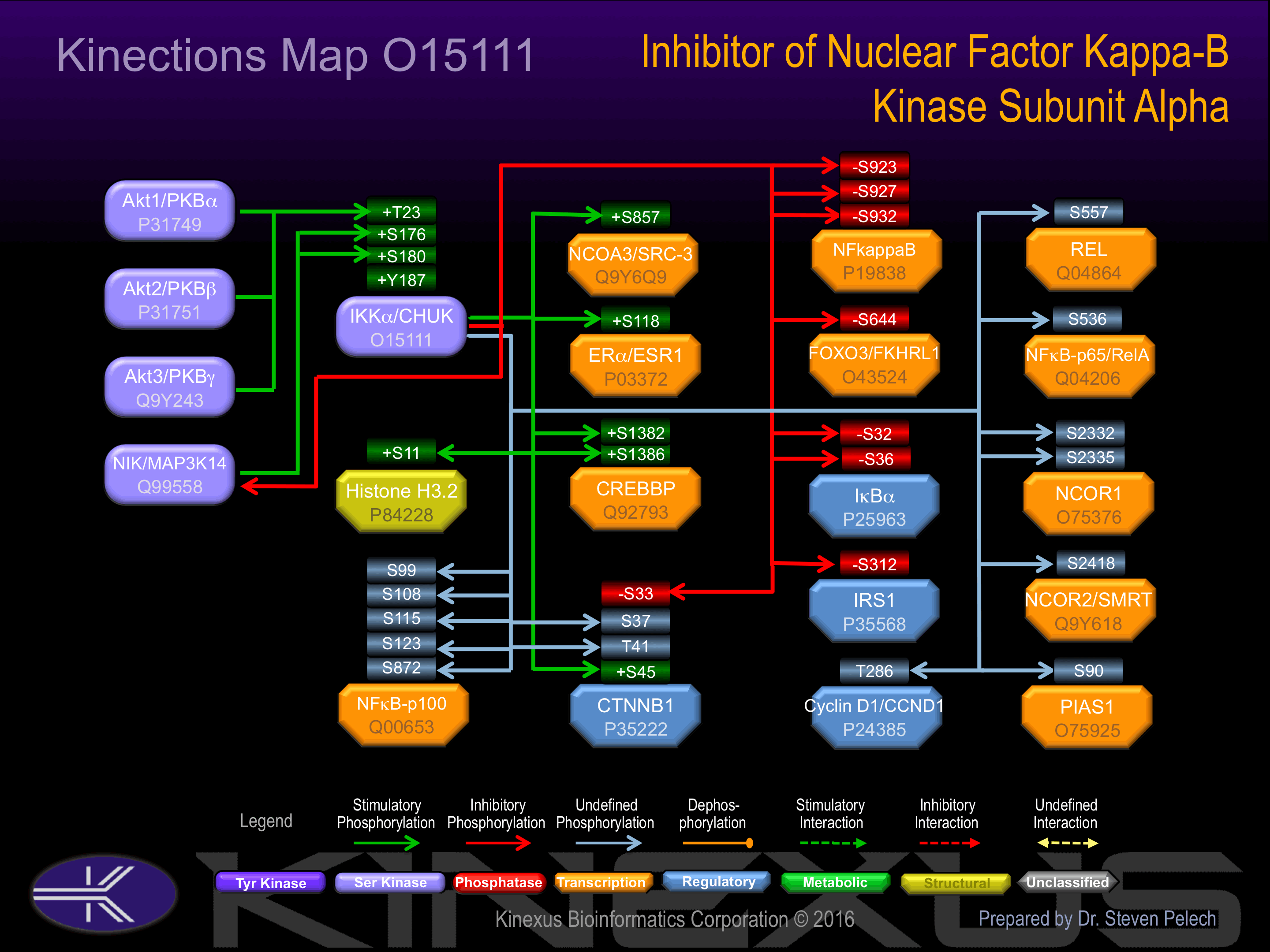
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| CBP (CREBBP) | Q92793 | S1382 | MKSRFVDSGEMSESF | + |
| CBP (CREBBP) | Q92793 | S1386 | FVDSGEMSESFPYRT | + |
| CTNNB1 | P35222 | S33 | QQQSYLDSGIHSGAT | - |
| CTNNB1 | P35222 | S37 | YLDSGIHSGATTTAP | ? |
| CTNNB1 | P35222 | S45 | GATTTAPSLSGKGNP | + |
| CTNNB1 | P35222 | T41 | GIHSGATTTAPSLSG | ? |
| Cyclin D1 (CCND1) | P24385 | T286 | EEVDLACTPTDVRDV | |
| ERa (ESR1) | P03372 | S118 | LHPPPQLSPFLQPHG | + |
| FOXO3 (FKHRL1) | O43524 | S644 | GLDFNFDSLISTQNV | - |
| H3.2 | P84228 | S11 | TKQTARKSTGGKAPR | + |
| IkBa | P25963 | S32 | LLDDRHDSGLDSMKD | - |
| IkBa | P25963 | S36 | RHDSGLDSMKDEEYE | - |
| IRS1 | P35568 | S312 | TESITATSPASMVGG | - |
| N-CoR1 | O75376 | S2332 | KLISKSNSRKSKSPI | |
| N-CoR1 | O75376 | S2335 | SKSNSRKSKSPIPGQ | |
| NCOA3 (SRC-3) | Q9Y6Q9 | S857 | PPYNRAVSLDSPVSV | + |
| NCOR2 | Q9Y618 | S2418 | AKVSGRPSSRKAKSP | |
| NFkB-p100 | Q00653 | S108 | PPRAHAHSLVGKQCS | |
| NFkB-p100 | Q00653 | S115 | SLVGKQCSELGICAV | |
| NFkB-p100 | Q00653 | S123 | ELGICAVSVGPKDMT | |
| NFkB-p100 | Q00653 | S872 | DSAYGSQSVEQEAEK | |
| NFkB-p100 | Q00653 | S99 | EVDLVTHSDPPRAHA | |
| NFkB-p105 | P19838 | S923 | DELRDSDSVCDSGVE | - |
| NFkB-p105 | P19838 | S927 | DSDSVCDSGVETSFR | - |
| NFkB-p105 | P19838 | S932 | CDSGVETSFRKLSFT | - |
| NFkB-p65 | Q04206 | S536 | SGDEDFSSIADMDFS | |
| PIAS1 | O75925 | S90 | SPMPATLSPSTIPQL | |
| REL | Q04864 | S557 | SNTTVFVSQSDAFEG |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
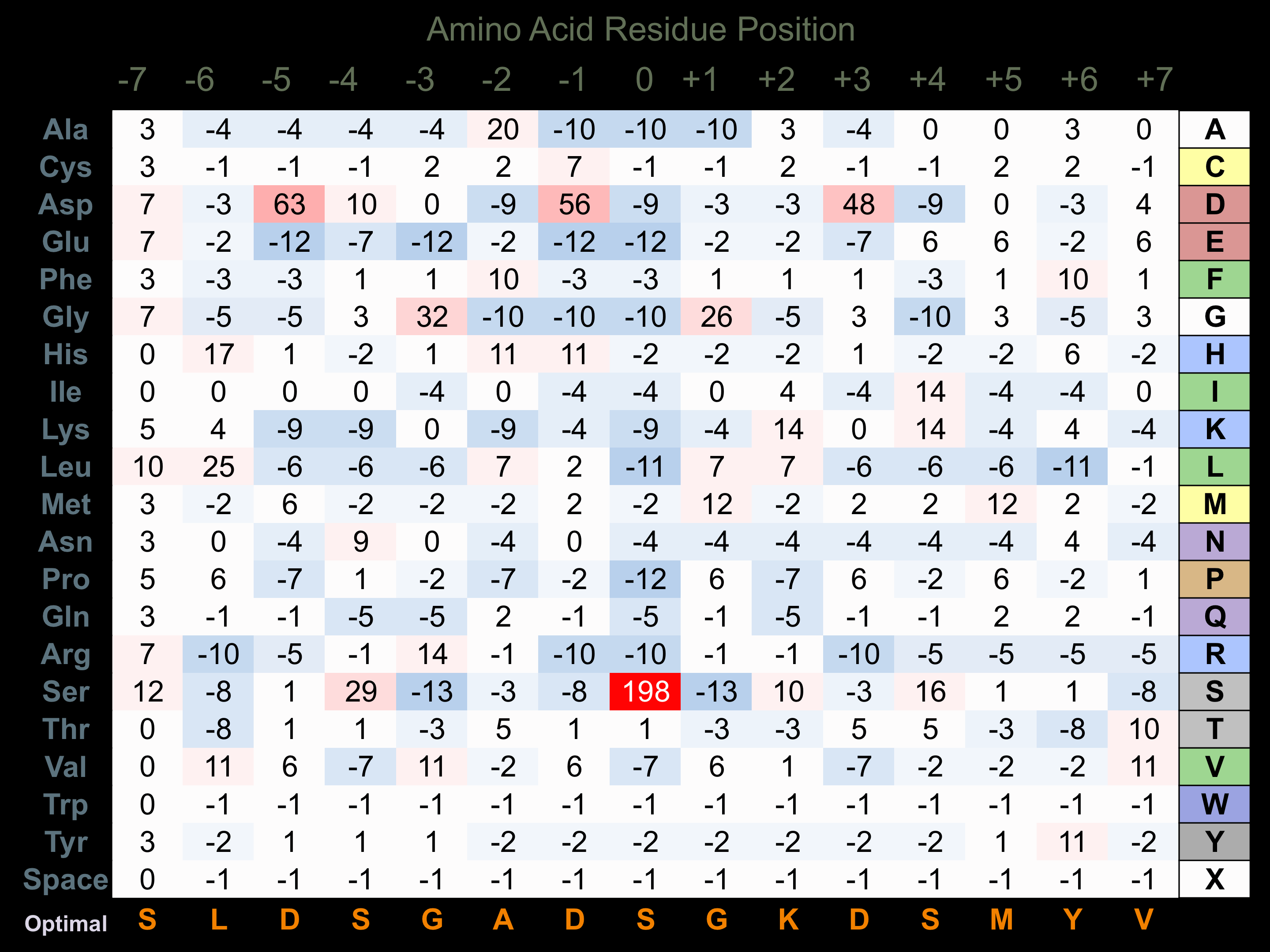
Matrix Type:
Experimentally derived from alignment of 32 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Gastrointestinal disorders and skin disorders
Specific Diseases (Non-cancerous):
Cocoon syndrome (COCOS); Skin papilloma
Comments:
IKKa gene mutations are associated with Cocoon syndrome, which is featured with multiple fetal malformations such as defective face and seemingly absent limbs.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +51, p<0.059); Breast epithelial carcinomas (%CFC= +110, p<0.064); and Ovary adenocarcinomas (%CFC= -49, p<0.003). The COSMIC website notes an up-regulated expression score for IKKa in diverse human cancers of 306, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 319 for this protein kinase in human cancers was 5.3-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25383 diverse cancer specimens. This rate is only -9 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.39 % in 1362 large intestine cancers tested; 0.24 % in 895 skin cancers tested; 0.2 % in 603 endometrium cancers tested; 0.15 % in 548 urinary tract cancers tested; 0.11 % in 127 biliary tract cancers tested; 0.1 % in 273 cervix cancers tested; 0.09 % in 589 stomach cancers tested; 0.09 % in 1768 lung cancers tested; 0.08 % in 958 upper aerodigestive tract cancers tested; 0.08 % in 1512 liver cancers tested; 0.06 % in 710 oesophagus cancers tested; 0.06 % in 238 bone cancers tested; 0.05 % in 1467 pancreas cancers tested; 0.04 % in 382 soft tissue cancers tested; 0.03 % in 891 ovary cancers tested; 0.03 % in 1546 breast cancers tested; 0.02 % in 558 thyroid cancers tested; 0.02 % in 2103 central nervous system cancers tested; 0.01 % in 939 prostate cancers tested; 0.01 % in 2009 haematopoietic and lymphoid cancers tested; 0.01 % in 1276 kidney cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: K237R (5).
Comments:
Only 4 deletions, 3 insertions, and no complex mutations are noted on the COSMIC website.