Nomenclature
Short Name:
RET
Full Name:
Proto-oncogene tyrosine-protein kinase receptor ret
Alias:
- CDHF12
- HSCR1
- MEN2A
- MEN2B
- MTC1
- PTC; RET51; RET-ELE1
- C-ret
- EC 2.7.10.1
- GDNF receptor
- Glial cell line-derived neurotropic factor receptor
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Ret
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
124,319
# Amino Acids:
1114
# mRNA Isoforms:
2
mRNA Isoforms:
124,319 Da (1114 AA; P07949); 119,847 Da (1072 AA; P07949-2)
4D Structure:
Phosphorylated form interacts with the PBT domain of DOK2, DOK4 and DOK5
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
N-GlcNAcylated:
N98, N151, N199, N336, N343, N361, N367, N377, N394, N448, N468, N554, .
Serine phosphorylated:
S159, S686, S688, S696+, S705, S891, S904, S1065, S1114.
Threonine phosphorylated:
T675, T754, T1111.
Tyrosine phosphorylated:
Y687, Y752, Y791, Y806+, Y809+, Y826, Y864+, Y900+, Y905+, Y928+, Y952+, Y981+, Y1015-, Y1029, Y1062+, Y1090, Y1096.
Ubiquitinated:
K1060, K1107.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1094
39
1404
 7
7
74
20
68
 2
2
25
38
26
 30
30
333
151
688
 49
49
540
33
453
 4
4
41
117
104
 27
27
300
52
442
 46
46
499
83
1084
 33
33
366
21
212
 5
5
55
162
63
 4
4
47
63
80
 45
45
487
303
611
 4
4
40
71
46
 4
4
46
13
45
 5
5
59
52
69
 5
5
52
23
31
 12
12
127
401
1471
 5
5
53
41
69
 6
6
71
155
62
 27
27
292
159
333
 6
6
64
54
64
 4
4
44
59
56
 3
3
34
44
43
 4
4
44
47
63
 4
4
47
50
48
 62
62
675
101
1694
 3
3
38
71
34
 4
4
42
45
54
 4
4
44
40
57
 20
20
224
42
159
 29
29
321
42
264
 57
57
621
45
549
 2
2
18
85
94
 70
70
761
83
714
 10
10
106
48
84
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 28.5
28.5
42.3
95 27.9
27.9
41.6
95 -
-
-
87 -
-
-
- 86.5
86.5
92.1
87 -
-
-
- 83.4
83.4
90.2
83.5 29
29
43.1
85 -
-
-
- 43
43
50.5
- 27.6
27.6
43.5
67.5 28.1
28.1
42.4
63 27.7
27.7
42
61.5 -
-
-
- 23.6
23.6
37.7
- -
-
-
- -
-
-
- 24.1
24.1
41.2
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | STAT3 - P40763 |
| 2 | DOK1 - Q99704 |
| 3 | DOK5 - Q9P104 |
| 4 | CRK - P46108 |
| 5 | CBL - P22681 |
| 6 | FRS2 - Q8WU20 |
| 7 | DOK2 - O60496 |
| 8 | AKAP5 - P24588 |
| 9 | PRKAR2A - P13861 |
| 10 | GRB7 - Q14451 |
| 11 | NRTN - Q99748 |
| 12 | PIK3R1 - P27986 |
| 13 | CBLB - Q13191 |
| 14 | GDNF - P39905 |
| 15 | FAU - P62861 |
Regulation
Activation:
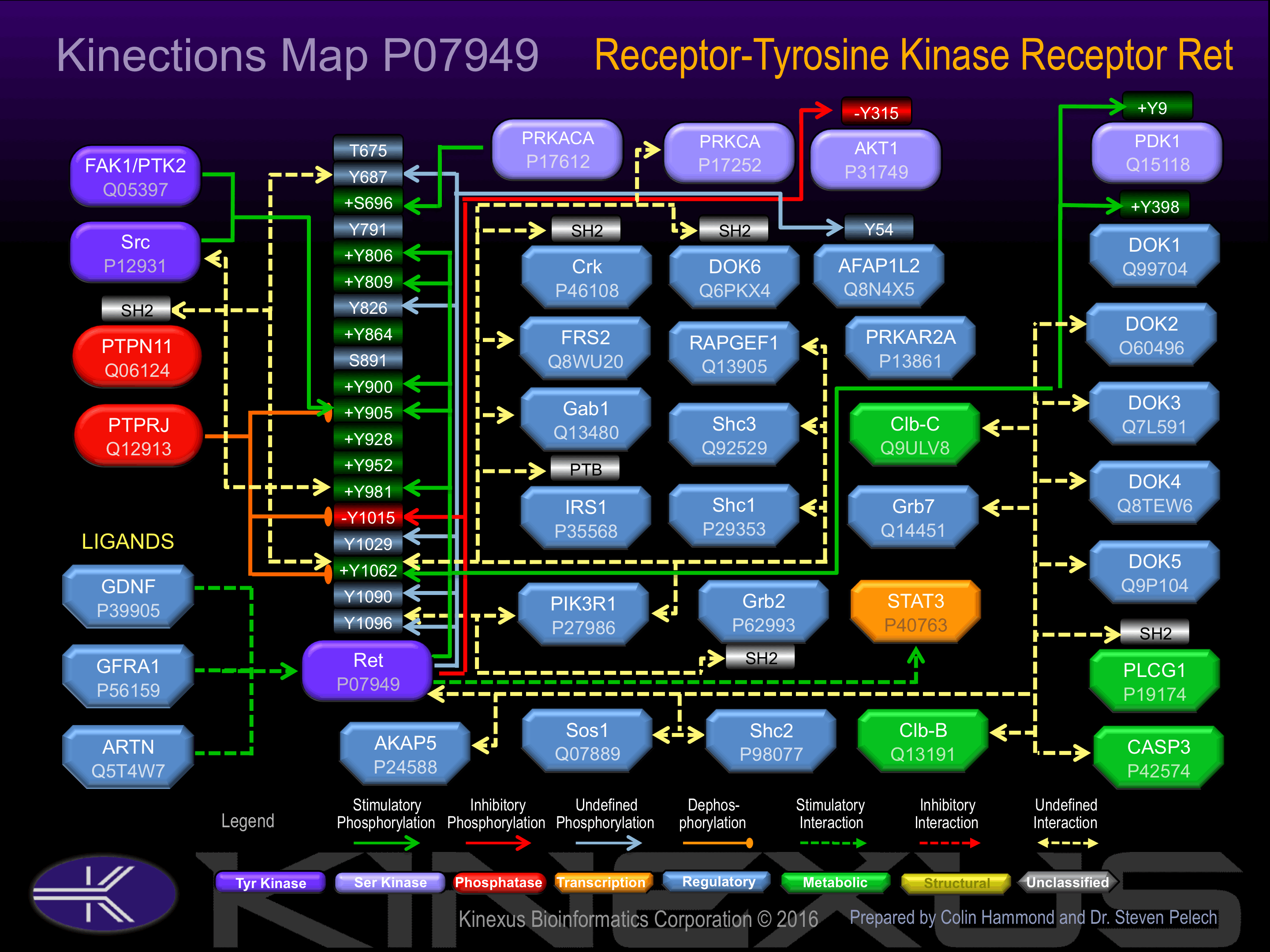
Activated by binding glial cell line-derived neurotropic factor, which induces dimerization and autophosphorylation. Phosphorylation of Ser-696, Tyr-806, Tyr-809, Tyr-864, Tyr-900, Tyr-905, Tyr-928 and Tyr-952 increases phosphotransferase activity. Phosphorylation of Tyr-981 increases phosphotransferase activity and induces interaction with Src Phosphorylation of Tyr-1062 increases phosphotransferase activity, and induces binding of Crk, Dok6, FRS2, Gab1, IRS1, PIK3R1, RapGEF1, Shc1 and Shc3. Phosphorylation of Tyr-1096 induces interaction with Grb2 and PIK3R1.
Inhibition:
Phosphorylation of Tyr-1015 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| RET | P07949 | Y687 | AQAFPVSYSSSGARR | ? |
| PKACa | P17612 | S696 | SSGARRPSLDSMENQ | + |
| RET | P07949 | Y806 | PLLLIVEYAKYGSLR | + |
| RET | P07949 | Y809 | LIVEYAKYGSLRGFL | + |
| RET | P07949 | Y826 | SRKVGPGYLGSGGSR | |
| RET | P07949 | Y900 | FGLSRDVYEEDSYVK | + |
| RET | P07949 | Y905 | DVYEEDSYVKRSQGR | + |
| SRC | P12931 | Y905 | DVYEEDSYVKRSQGR | + |
| FAK | Q05397 | Y905 | DVYEEDSYVKRSQGR | + |
| RET | P07949 | Y981 | DNCSEEMYRLMLQCW | + |
| RET | P07949 | Y1015 | MMVKRRDYLDLAAST | - |
| RET | P07949 | Y1029 | TPSDSLIYDDGLSEE | |
| RET | P07949 | Y1062 | TWIENKLYGMSDPNW | + |
| RET | P07949 | Y1090 | TNTGFPRYPNDSVYA | |
| RET | P07949 | Y1096 | RYPNDSVYANWMLSP |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| AFAP1L2 | Q8N4X5 | Y54 | SSSSDEEYIYMNKVT | |
| Akt1 (PKBa) | P31749 | Y315 | TFCGTPEYLAPEVLE | - |
| Dok1 p62 | Q99704 | Y398 | ARVKEEGYELPYNPA | + |
| PDK1 (PDPK1) | O15530 | Y9 | ARTTSQLYDAVPIQS | + |
| Ret | P07949 | Y1015 | MMVKRRDYLDLAAST | - |
| Ret | P07949 | Y1029 | TPSDSLIYDDGLSEE | |
| Ret | P07949 | Y1062 | TWIENKLYGMSDPNW | + |
| Ret | P07949 | Y1090 | TNTGFPRYPNDSVYA | |
| Ret | P07949 | Y1096 | RYPNDSVYANWMLSP | |
| Ret | P07949 | Y687 | AQAFPVSYSSSGARR | ? |
| Ret | P07949 | Y806 | PLLLIVEYAKYGSLR | + |
| Ret | P07949 | Y809 | LIVEYAKYGSLRGFL | + |
| Ret | P07949 | Y826 | SRKVGPGYLGSGGSR | |
| Ret | P07949 | Y900 | FGLSRDVYEEDSYVK | + |
| Ret | P07949 | Y905 | DVYEEDSYVKRSQGR | + |
| Ret | P07949 | Y981 | DNCSEEMYRLMLQCW | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
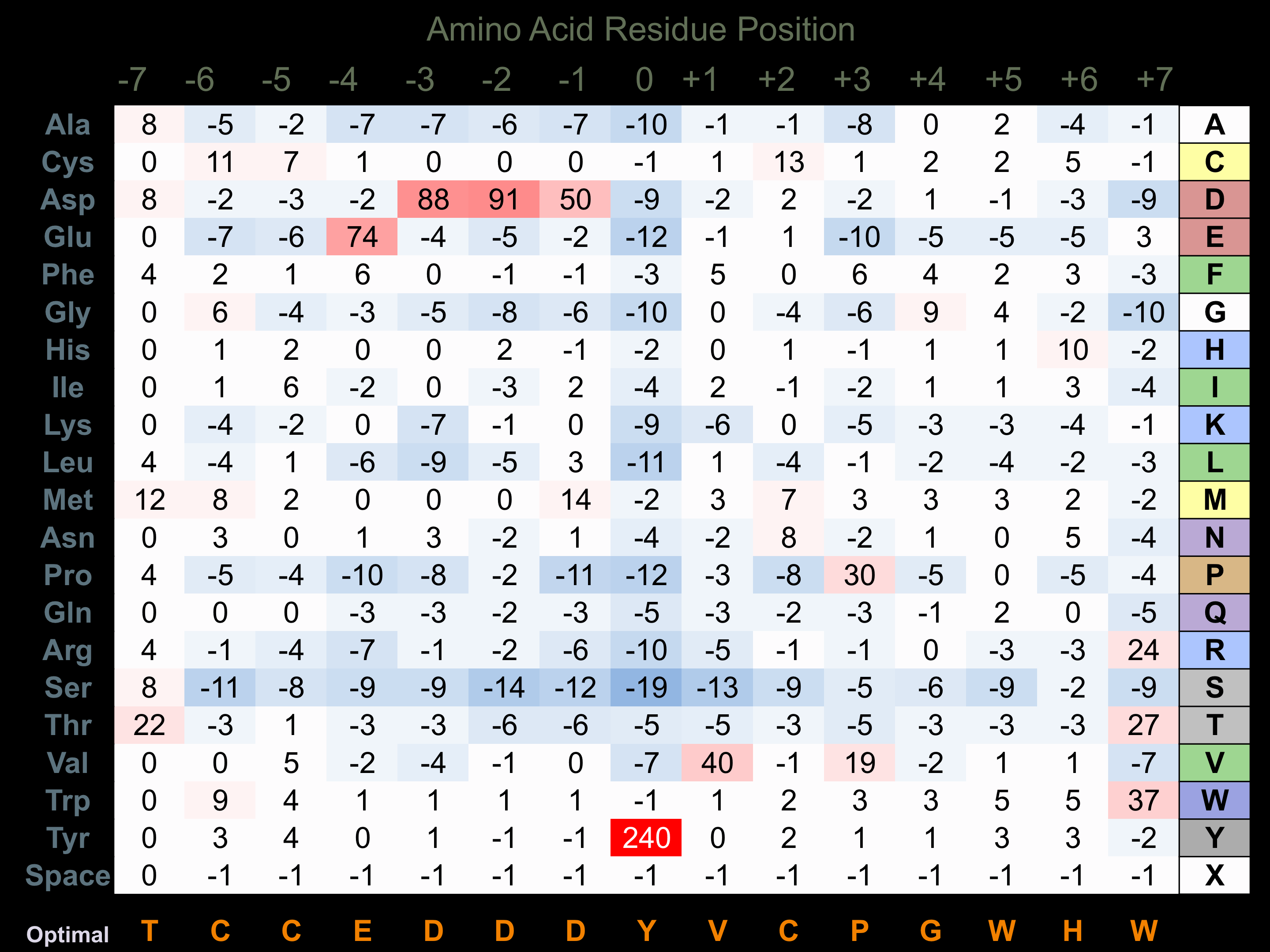
Matrix Type:
Experimentally derived from alignment of 19 known protein substrate phosphosites and 57 peptides phosphorylated by recombinant Ret in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, gastrointestinal, nephrological, endocrine, eye, ear, cardiovascular, and skin disorders
Specific Diseases (Non-cancerous):
Hirschsprung's disease; Ret-related Hirschsprung disease; Ret-related Pheochromocytoma; Ret-related renal Adysplasia; Renal agenesis; Congenital central hypoventilation syndrome; Renal hypoplasia, Isolated; renal hypodysplasia/aplasia 1; Pheochromocytoma, modifier of; Central hypoventilation syndrome; Congenital, with or without Hirschsprung disease; Wagenmann-Froboese syndrome; Hyperparathyroidism; Neurofibromatosis; Von Hippel-Lindau disease; Cowden disease; Unilateral renal agenesis; Hypoganglionosis; Lichen amyloidosis; Medullary sponge kidney; Hereditary paraganglioma-pheochromocytoma syndromes; Intestinal obstruction; Familial Isolated hyperparathyroidism; Primary cutaneous amyloidosis; Renal dysplasia; Struma ovarii; Parathyroid gland disease; Sipple syndrome; Thyroiditis
Comments:
Mutations of this gene are associated with Hirschsprung disease 1, which is characterized by absence of enteric ganglia along a variable length of the intestine. R67H, R114H, A432E and P1039L substitutions are involved in congenital central hypoventilation syndrome, that often shows abnormal control of respiration in the absence of neuromuscular or lung disease, or an identifiable brain stem lesion. It may also associate with renal abnormalities.
Specific Cancer Types:
Multiple endocrine neoplasia (MEN); Pheochromocytomas (PCC); Thyroid cancer (TC); Multiple endocrine neoplasias (MEN) type 2a; Thyroid medullary carcinomas (MTC); Papillary carcinomas (PTC); Familial medullary thyroid carcinomas (FMTC); Paraganglioma (PGL); Papillary thyroid carcinomas (PTC); Follicular adenoma (FA); Follicular thyroid carcinomas (FTC); Primary peritoneal carcinomas (PPC); Pancreatic islet cell tumours; Sacrococcygeal teratoma (SCT); Peritoneal carcinomas; Familial papillary thyroid carcinomas (fPTC); neuroblastomas (NB)
Comments:
RET is a known oncoprotein (OP). Cancer-related mutations in human tumours point to a gain of function of the protein kinase. The active form of the protein kinase normally acts to promote tumour cell proliferation. Mutations in the Ret gene are associated with the disorders multiple endocrine neoplasia, type IIA, multiple endocrine neoplasia, type IIB, and medullary thyroid carcinoma. Ret was identified as a potential low-penetrance gene for apparently sporadic pheochromocytoma.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= -48, p<0.006); Brain glioblastomas (%CFC= -92, p<0.048); Breast epithelial carcinomas (%CFC= +253, p<0.068); Breast non-basal-like cancer (BLC) (%CFC= +49, p<(0.0003); Skin fibrosarcomas (%CFC= +468, p<0.003); and Skin melanomas (%CFC= -51, p<0.048). The COSMIC website notes an up-regulated expression score for RET in diverse human cancers of 348, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.28 % in 34161 diverse cancer specimens. This rate is 3.75-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 2.29 % in 2675 thyroid cancers tested; 0.5 % in 1528 large intestine cancers tested; 0.3 % in 1378 skin cancers tested; 0.23 % in 856 stomach cancers tested; 0.22 % in 708 endometrium cancers tested; 0.22 % in 534 urinary tract cancers tested; 0.18 % in 2555 lung cancers tested; 0.13 % in 696 oesophagus cancers tested; 0.07 % in 1316 kidney cancers tested; 0.05 % in 2832 breast cancers tested; 0.05 % in 2130 haematopoietic and lymphoid cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: M918T (496); C634R (33); C634Y (19); C634W (16).
Comments:
In human tumours, there are two main regions of point mutations: one small cluster at amino acid residues 591-634 and a slightly larger cluster around M918 with many point mutations. Over 30 deletions (15 at E632 and 10 at D898), 2 (1 at C634) insertions and over 20 complex mutations (17 at E632) are also noted on the COSMIC website.