Nomenclature
Short Name:
IGF1R
Full Name:
Insulin-like growth factor I receptor
Alias:
- CD221
- MGC142170
- MGC142172
- EC 2.7.10.1
- IGFIR
- Insulin-like growth factor I receptor
- JTK13
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
InsR
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
154,793
# Amino Acids:
1367
# mRNA Isoforms:
1
mRNA Isoforms:
154,793 Da (1367 AA; P08069)
4D Structure:
Tetramer of 2 alpha and 2 beta chains linked by disulfide bonds. The alpha chains contribute to the formation of the ligand-binding domain, while the beta chain carries the kinase domain. Interacts with PIK3R1 and with the PTB/PID domains of IRS1 and SHC1 in vitro when autophosphorylated on tyrosine residues. Forms a hybrid receptor with INSR, the hybrid is a tetramer consisting of 1 alpha chain and 1 beta chain of INSR and 1 alpha chain and 1 beta chain of IGF1R. Interacts with ARRB1 and ARRB2. Interacts with GRB10.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
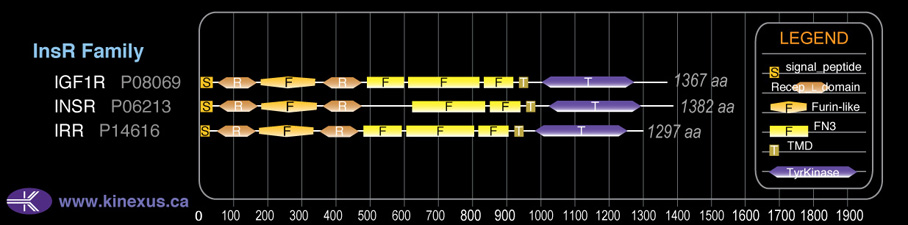
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 1 | 30 | signal_peptide |
| 51 | 161 | Recep_L_domain |
| 175 | 333 | Furin-like |
| 352 | 467 | Recep_L_domain |
| 488 | 606 | FN3 type 1 |
| 611 | 689 | FN3 type 2 |
| 831 | 926 | FN3 type 3 |
| 936 | 958 | TMD |
| 999 | 1266 | TyrKc |
| 999 | 1274 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Methylated:
R471, R480, K1354.
N-GlcNAcylated:
N51, N102, N135, N314, N417, N438, N534, N607, N622, N640, N747, N756, N764, N900, N913.
Other:
Glycyl lysine isopeptide (Lys-Gly) (interchain with G-Cter in ubiquitin) linked to K1168, K1171.
Serine phosphorylated:
S3, S5, S9, S12, S29, S623, S624, S982, S1193, S1278-, S1310+, S1311+, S1312+, S1313+, S1321.
Sumoylated:
K1055, K1130, K1150.
Threonine phosphorylated:
T11, T28, T123, T1163+, T1366.
Tyrosine phosphorylated:
Y113, Y115, Y131, Y577, Y580, Y973, Y980+, Y1014, Y1161+, Y1165+, Y1166+, Y1280, Y1281, Y1346+.
Ubiquitinated:
K1033.
Acetylated:
K1088.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 70
70
1013
57
1031
 10
10
146
23
139
 15
15
219
23
181
 35
35
503
220
936
 56
56
809
56
688
 8
8
111
144
158
 16
16
237
71
399
 74
74
1076
91
2135
 23
23
338
24
284
 14
14
200
199
214
 2
2
36
58
41
 44
44
632
281
646
 4
4
62
56
69
 3
3
49
17
26
 10
10
145
49
176
 8
8
113
33
89
 17
17
252
558
2230
 10
10
141
34
128
 5
5
77
179
87
 45
45
647
218
679
 7
7
106
46
120
 7
7
98
52
98
 10
10
138
34
131
 10
10
141
34
125
 8
8
120
46
122
 100
100
1445
141
2633
 5
5
68
59
72
 9
9
131
34
114
 13
13
182
34
174
 9
9
126
70
97
 27
27
385
30
404
 31
31
449
67
700
 7
7
98
143
205
 50
50
716
135
630
 6
6
82
83
86
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.7
99.7
99.7
100 99.5
99.5
99.6
100 -
-
-
98 -
-
-
98 98.2
98.2
98.8
98 -
-
-
- 95.6
95.6
97.8
96 95.7
95.7
98
96 -
-
-
- -
-
-
- 21.3
21.3
35.5
85 75.5
75.5
85.9
77 66.6
66.6
79.2
69 -
-
-
- 26.8
26.8
40.3
42 -
-
-
- 27
27
41.5
36 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
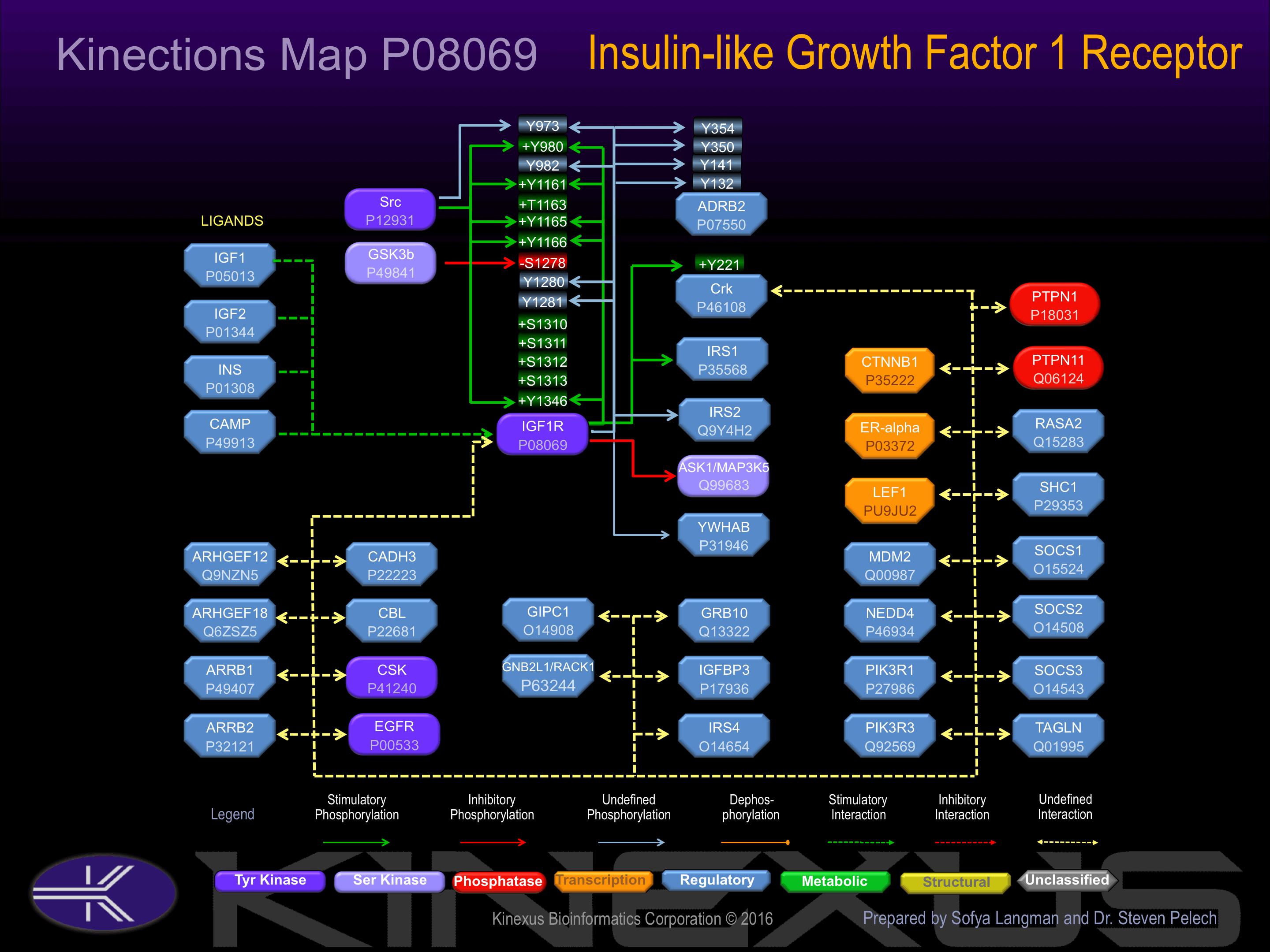
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | IRS1 - P35568 |
| 2 | GRB10 - Q13322 |
| 3 | SHC1 - P29353 |
| 4 | IGF1 - P01343 |
| 5 | PTPN1 - P18031 |
| 6 | IGF1 - P05019 |
| 7 | PTPN11 - Q06124 |
| 8 | YWHAB - P31946 |
| 9 | INS - P01308 |
| 10 | PIK3R1 - P27986 |
| 11 | ESR1 - P03372 |
| 12 | SOCS2 - O14508 |
| 13 | SOCS1 - O15524 |
| 14 | YWHAE - P62258 |
| 15 | PIK3R3 - Q92569 |
Regulation
Activation:
Activated by binding insulin-like growth factor I (IGF1), which induces the pre-dimerization receptor to autophosphorylation.Phosphorylation of Tyr-1161 and Tyr-1346 increases its phosphotransferase activity and induces interaction with Src. Phosphorylation of Tyr-980, Tyr-1161, Tyr-1165 and Tyr-1166 increases its phosphotransferase activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| SRC | P12931 | Y973 | RLGNGVLYASVNPEY | |
| IGF1R | P08069 | Y973 | RLGNGVLYASVNPEY | |
| SRC | P12931 | Y980 | YASVNPEYFSAADVY | + |
| IGF1R | P08069 | Y980 | YASVNPEYFSAADVY | + |
| IGF1R | P08069 | S982 | SVNPEYFSAADVYVP | |
| SRC | P12931 | Y1161 | FGMTRDIYETDYYRK | + |
| IGF1R | P08069 | Y1161 | FGMTRDIYETDYYRK | + |
| SRC | P12931 | Y1165 | RDIYETDYYRKGGKG | + |
| IGF1R | P08069 | Y1165 | RDIYETDYYRKGGKG | + |
| SRC | P12931 | Y1166 | DIYETDYYRKGGKGL | + |
| IGF1R | P08069 | Y1166 | DIYETDYYRKGGKGL | + |
| GSK3B | P49841 | S1278 | EPGFREVSFYYSEEN | - |
| IGF1R | P08069 | Y1280 | GFREVSFYYSEENKL | ? |
| IGF1R | P08069 | Y1281 | FREVSFYYSEENKLP | ? |
| SRC | P12931 | Y1346 | SFDERQPYAHMNGGR | + |
| IGF1R | P08069 | Y1346 | SFDERQPYAHMNGGR | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ADRB2 | P07550 | Y132 | CVIAVDRYFAITSPF | |
| ADRB2 | P07550 | Y141 | AITSPFKYQSLLTKN | |
| ADRB2 | P07550 | Y350 | RRSSLKAYGNGYSSN | |
| ADRB2 | P07550 | Y354 | LKAYGNGYSSNGNTG | |
| Crk | P46108 | Y221 | GGPEPGPYAQPSVNT | + |
| IGF1R | P08069 | S982 | SVNPEYFSAADVYVP | |
| IGF1R | P08069 | Y1161 | FGMTRDIYETDYYRK | + |
| IGF1R | P08069 | Y1165 | RDIYETDYYRKGGKG | + |
| IGF1R | P08069 | Y1166 | DIYETDYYRKGGKGL | + |
| IGF1R | P08069 | Y1280 | GFREVSFYYSEENKL | ? |
| IGF1R | P08069 | Y1281 | FREVSFYYSEENKLP | ? |
| IGF1R | P08069 | Y1346 | SFDERQPYAHMNGGR | + |
| IGF1R | P08069 | Y973 | RLGNGVLYASVNPEY | |
| IGF1R | P08069 | Y980 | YASVNPEYFSAADVY | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
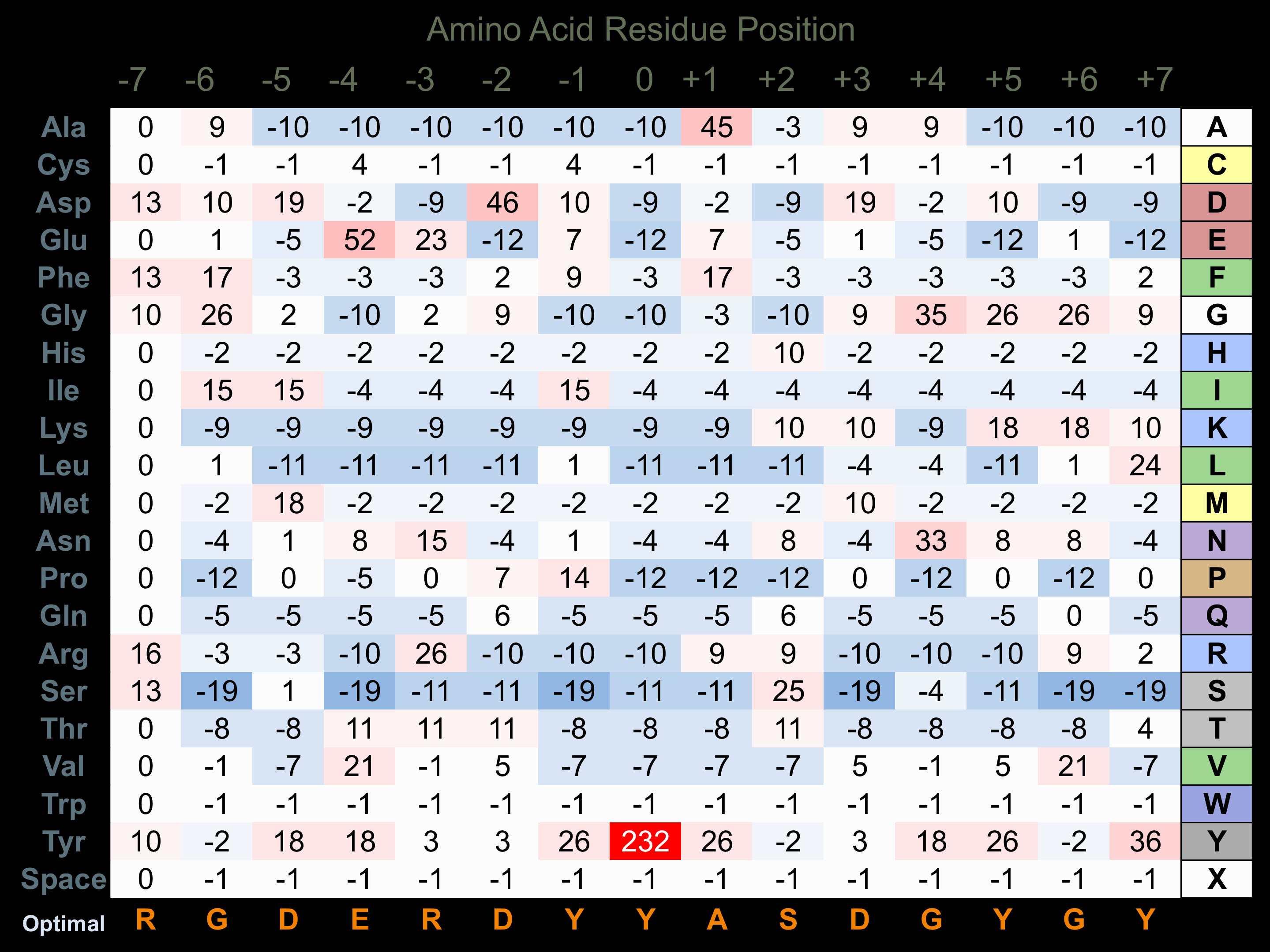
Matrix Type:
Experimentally derived from alignment of 23 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, endocrine disorders
Specific Diseases (Non-cancerous):
Insulin resistance; Insulin-like growth factor I (IGF1) deficiency; Lymphangioleiomyomatosis; Ring chromosome 15; Donohue syndrome; Rabson-Mendenhall syndrome; Insulin-like growth factor 1, resistance to
Comments:
Insulin-Like Growth Factor 1 Resistance (IGF1RES) to is a rare condition with resistance to IGF-1, and is related to Alzheimer’s and neuronitis. It is characterized by intrauterine growth retardation, poor postnatal growth, and increased plasma IGF1 levels. The mutations in IGF1RES, R138Q or K145N, leads to decreased IGF1R function. A R739Q mutation in IGF1RES impairs the production of mature IGF1R from the proreceptor. Several other mutations have been observed in the IGFR1 gene in patients with insulin-resistance, including a R108Q substitution mutation, a K115N substitution mutation, a R59X termination mutation, a R709Q substitution mutation, and a R481Q substitution mutation. These mutations are predicted to produce a loss of phosphotransferase activity of the IGFR1 gene. Therefore, loss-of-function of IGFR1 function appears to play a role in the pathogenesis of insulin resistance. Mutations in IGFR1 and abnormal IGF1 signalling are prime candidates for the cause of low birth weight, insulin resistance, and Type 2 diabetes. In animal studies, mice lacking IGFR1 expression in the beta cells of the pancreas displayed normal growth and development of beta cells, but had a reduction in GLUT2 (glucose transporter) and GCK (encodes glucokinase) expression, such that glucose-stimulated insulin release was impaired. In addition, mice lacking IGFR1 in all cells of their body died at birth due to respiratory failure and had severe growth retardation (~45% of expected size). In addition there was generalized organ hypoplasia, including skeletal muscles, central nervous system, and epidermis, as well as delayed ossificiation of the bones. By contrast, down-regulation of the IGF1 pathway or IGF1 levels in plasma has been shown to associate with an increased life span. >Insulin resistance is a disease characterized by a failure of cells to respond to normal levels of circulating insulin, resulting in hyperglycemia due to impaired glucose uptake and improper glycogen management. This disease is commonly associated with an overproduction of insulin by the beta cells in the pancreas, which leads to hyperinsulinemia, and can contribute to the development of Type 2 diabetes or latent autoimmune diabetes in adults. >Lymphangioleiomyomatosis (LAM) is a rare lung disease characterized by uncontrolled cell growth throughout the body, including the lungs, lymphatic system, and kidneys. LAM also generates cysts from the continued growth of cell masses in the lung, destroying tissue and causing alveoli damage, resulting in obstruction and impaired oxygen movement and availability. >The fatal Donohue syndrome (DS), also known as Leprechanumism, is an extremely rare genetic disease characterized by stunted growth, and elfin features such as protruding ears, flaring nostrils, and thick lips. DS can be caused by congenital mutations in the insulin receptor (IR). >Rabson-Mendenhall syndrome is another rare genetic disease characterized by a severe resistance to insulin. Symptoms of the disease include impaired intrauterine and post-natal growth, hypertrophy of muscle and fat tissues, and craniofacial, dental, skin and nail abnormalities. >Ring Chromosome 15 (R15) is related to Down syndrome and infertility, but can have highly variable symptoms. >Insulin-Like Growth Factor I Deficiency is the low production of IGF1. The disorder is characterized by cognitive impairment, short hand (short digits), and pectus excavatum (sunken ribs near the sternum, causing a sunken chest).
Specific Cancer Types:
Breast cancer; Colorectal cancer; Lung cancer (LC)
Comments:
IGF1R appears to be an oncoprotein (OP). Cancer-related mutations in human tumours point to a gain of function of the protein kinase. The active form of the protein kinase normally acts to promote tumour cell proliferation. IGF1R been shown to be an important signalling component in several types of human cancers. In particular, the activated receptor is critical for tumour transformation, malignant cell survival, and the maintenance of the transformed phenotype. In addition, activated IGF1R protein functions to inhibit apoptosis via indirect phosphorylation of the BAD protein via Akt, as well as plays an important role in the regulation of cancer cell motility and metastasis. Several cancer types display increased expression of IGF1R and/or increased catalytic activity of the protein, including prostate cancer, colorectal adenoma, and melanoma. Additionally, cross-talk between the IGF signalling pathway and the estrogen receptor signalling pathway has been demonstrated in breast cancer cells as a central mechanism of cell growth regulation. Furthermore, therapies aimed at the inhibition of IGF1R signalling have shown promise as potential cancer treatments. For example, different classes of molecules have been tested as potential cancer therapeutics, including antisense RNA, monoclonal antibodies, and dominant-negative IGF1R protein variants. These molecules have been demonstrated to reverse the transformed phenotype of cancer cells in both human and rodent cancer cell lines.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= +87, p<0.0001); Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= +100, p<0.093); Cervical cancer stage 1B (%CFC= +100, p<0.007); Cervical cancer stage 2A (%CFC= +97, p<0.033); Cervical cancer stage 2B (%CFC= +92, p<0.091); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +217, p<0.102); Prostate cancer - primary (%CFC= +83, p<0.0001); Skin melanomas - malignant (%CFC= +142, p<0.0001); and Uterine fibroids (%CFC= +61, p<0.078). The COSMIC website notes an up-regulated expression score for IGF1R in diverse human cancers of 478, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 10 for this protein kinase in human cancers was 0.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis. Tyrosine phosphorylation and signalling can be reduced with an Y980F mutation, which results in inhibited IRS1 or SHC1 interactions with IGF1R. However, this mutation does not inhibit interactions with PIK3R1 or GRB10. The IGF1R phosphotransferase activity can be inhibited with a K1033A mutation, which inhibits interactions between IGF1R and IRS1, SHC1, and PIK3R1. A Y1346F mutation can prevent GRB10 from binding to IGF1R.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25698 diverse cancer specimens. This rate is very similar (+ 2% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.39 % in 1128 large intestine cancers tested; 0.35 % in 590 stomach cancers tested; 0.21 % in 807 skin cancers tested; 0.19 % in 602 endometrium cancers tested; 0.1 % in 1270 liver cancers tested; 0.08 % in 1946 lung cancers tested; 0.06 % in 1397 kidney cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,917 cancer specimens
Comments:
Only 5 deletions, no insertions or complex mutations are noted on the COSMIC website.