Nomenclature
Short Name:
MET
Full Name:
Hepatocyte growth factor receptor
Alias:
- C-met
- EC 2.7.10.1
- Kinase Met
- Met proto- oncogene tyrosine kinase
- Met proto-oncogene
- RCCP2
- Hepatocyte growth factor receptor
- HGF receptor
- HGFR
- HGF-SF receptor
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Met
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
155,541
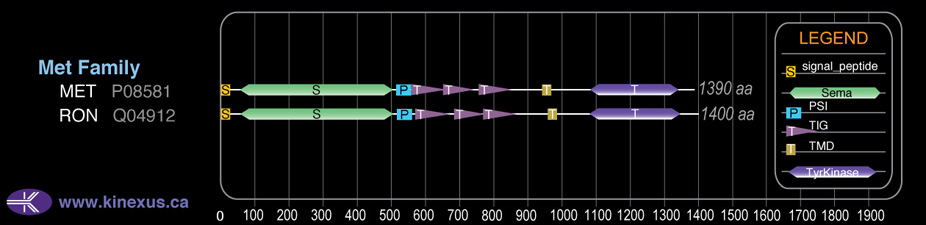
# Amino Acids:
1390
# mRNA Isoforms:
3
mRNA Isoforms:
157,712 Da (1408 AA; P08581-2); 155,541 Da (1390 AA; P08581); 85,745 Da (764 AA; P08581-3)
4D Structure:
Heterodimer formed of an alpha chain (50 kDa) and a beta chain (145 kDa) which are disulfide linked. Binds PLXNB1 and GRB2. Interacts with SPSB1, SPSB2 and SPSB4 By similarity. Interacts with INPP5D/SHIP1. When phosphorylated at Tyr-1356, interacts with INPPL1/SHIP2. Interacts with RANBP9 and RANBP10, as well as SPSB1, SPSB2, SPSB3 and SPSB4. SPSB1 binding occurs in the presence and in the absence of HGF, however HGF treatment has a positive effect on this interaction. Interacts with MUC20; prevents interaction with GRB2 and suppresses hepatocyte growth factor-induced cell proliferation. Interacts with GRB10.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K1190.
N-GlcNAcylated:
N45, N106, N149, N202, N399, N405, N607, N635, N785, N879, N930.
Serine phosphorylated:
S637, S641, S755, S758, S966, S985-, S988, S990, S997, S1000, S1016, S1020, S1236+, S1342, S1367, S1370, S1371.
Threonine phosphorylated:
T17, T750, T759, T977, T992, T993, T1006, T1096, T1241+, T1289, T1343, T1355.
Tyrosine phosphorylated:
Y522, Y745, Y830, Y971, Y1003+, Y1026, Y1093, Y1159, Y1194, Y1230+, Y1234+, Y1235+, Y1307, Y1313, Y1349-, Y1356-, Y1365.
Ubiquitinated:
K189, K223, K324, K765, K962, K1104, K1161, K1190, K1193, K1219, K1232, K1240, K1318.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 55
55
1096
55
1238
 2
2
43
26
54
 4
4
82
4
46
 37
37
736
172
1753
 30
30
597
47
550
 5
5
107
158
210
 17
17
343
67
486
 100
100
1990
69
3231
 19
19
378
31
360
 6
6
127
168
95
 5
5
103
45
85
 33
33
660
338
675
 3
3
50
48
27
 2
2
47
21
41
 3
3
62
35
57
 2
2
33
31
31
 2
2
37
444
35
 3
3
62
22
49
 6
6
122
163
83
 25
25
492
215
588
 2
2
47
30
54
 2
2
48
39
40
 4
4
83
8
63
 3
3
62
20
66
 2
2
46
30
50
 76
76
1513
101
2385
 2
2
43
51
34
 4
4
89
20
74
 5
5
94
20
126
 9
9
185
56
163
 16
16
322
36
444
 40
40
794
66
1241
 69
69
1377
106
2095
 39
39
780
104
691
 9
9
171
61
172
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.8
99.8
99.9
100 99.8
99.8
99.9
99 -
-
-
92 -
-
-
90 98.1
98.1
98.7
91 35.2
35.2
52.3
- 88.8
88.8
94.5
90 87.8
87.8
93.6
89 87.8
87.8
93.6
- 77.3
77.3
86.7
- 77.3
77.3
86.7
73 -
-
-
64 -
-
-
52 -
-
-
- 48.4
48.4
66
- -
-
-
- -
-
-
- -
-
-
- 28.7
28.7
49
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | HGF - P14210 |
| 2 | GAB1 - Q13480 |
| 3 | PTPN1 - P18031 |
| 4 | PTPN11 - Q06124 |
| 5 | CBL - P22681 |
| 6 | CASP3 - P42574 |
| 7 | SPSB1 - Q96BD6 |
| 8 | GLMN - Q92990 |
| 9 | GRB2 - P62993 |
| 10 | MUC20 - Q8N307 |
| 11 | FAS - P25445 |
| 12 | SRC - P12931 |
| 13 | CTNNB1 - P35222 |
| 14 | STAT3 - P40763 |
| 15 | BAG1 - Q99933 |
Regulation
Activation:
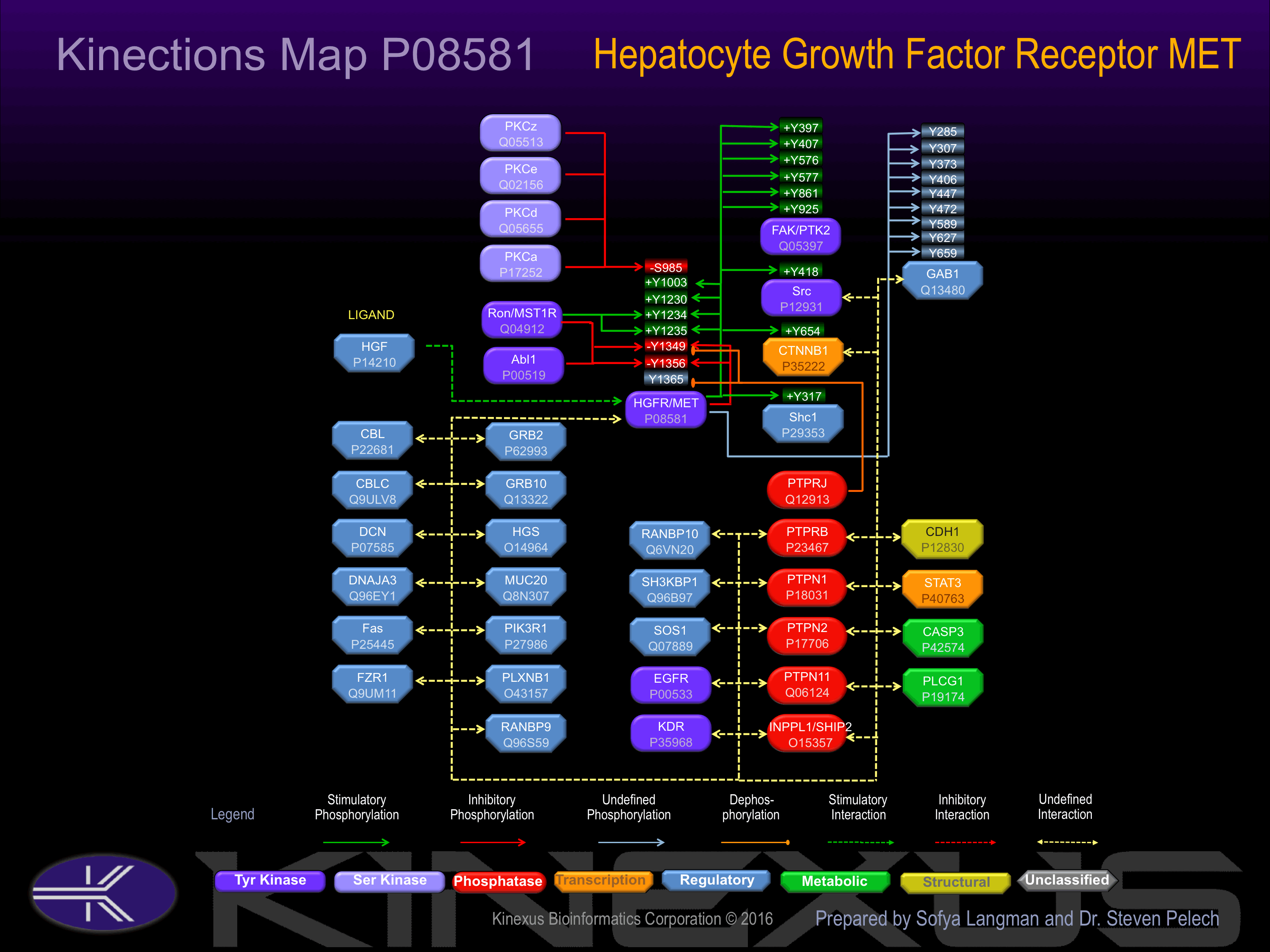
Activated by binding hepatocyte growth factor (HGF), which induces dimerization and autophosphorylation. Phosphorylation of Tyr-1003 increases phosphotransferase activity and induces interaction with Cbl. Phosphorylation of Tyr-1230 and Tyr-1235 increases phosphotransferase activity. Phosphorylation of Tyr-1234 increases phosphotransferase activity and induces interaction with Grb2.
Inhibition:
Phosphorylation of Ser-985 inhibits phosphotransferase activity. Phosphorylation of Tyr-1349 inhibits phosphotransferase activity, but induces interaction with FAK, Gab1, Grb2, Met, PIK3R1, PLCG1 and Src.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| PKCz | Q05513 | S985 | PHLDRLVSARSVSPT | - |
| PKCa | P17252 | S985 | PHLDRLVSARSVSPT | - |
| PKCe | Q02156 | S985 | PHLDRLVSARSVSPT | - |
| PKCd | Q05655 | S985 | PHLDRLVSARSVSPT | - |
| MET | P08581 | Y1003 | VSNESVDYRATFPED | + |
| MET | P08581 | Y1230 | FGLARDMYDKEYYSV | + |
| MET | P08581 | Y1234 | RDMYDKEYYSVHNKT | + |
| RON | Q04912 | Y1234 | RDMYDKEYYSVHNKT | + |
| MET | P08581 | Y1235 | DMYDKEYYSVHNKTG | + |
| RON | Q04912 | Y1235 | DMYDKEYYSVHNKTG | + |
| ABL | P00519 | Y1349 | STFIGEHYVHVNATY | - |
| MET | P08581 | Y1349 | STFIGEHYVHVNATY | - |
| RON | Q04912 | Y1349 | STFIGEHYVHVNATY | - |
| ABL | P00519 | Y1356 | YVHVNATYVNVKCVA | - |
| MET | P08581 | Y1356 | YVHVNATYVNVKCVA | - |
| RON | Q04912 | Y1356 | YVHVNATYVNVKCVA | - |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| FAK (PTK2) | Q05397 | Y397 | SVSETDDYAEIIDEE | + |
| FAK (PTK2) | Q05397 | Y407 | IIDEEDTYTMPSTRD | + |
| FAK (PTK2) | Q05397 | Y576 | RYMEDSTYYKASKGK | + |
| FAK (PTK2) | Q05397 | Y577 | YMEDSTYYKASKGKL | + |
| FAK (PTK2) | Q05397 | Y861 | PIGNQHIYQPVGKPD | + |
| FAK (PTK2) | Q05397 | Y925 | DRSNDKVYENVTGLV | + |
| GAB1 | Q13480 | Y285 | TEADGELYVFNTPSG | |
| GAB1 | Q13480 | Y307 | MRHVSISYDIPPTPG | |
| GAB1 | Q13480 | Y373 | ASDTDSSYCIPTAGM | |
| GAB1 | Q13480 | Y406 | DASSQDCYDIPRAFP | |
| GAB1 | Q13480 | Y447 | SEELDENYVPMNPNS | |
| GAB1 | Q13480 | Y472 | EPIQEANYVPMTPGT | |
| GAB1 | Q13480 | Y589 | SHDSEENYVPMNPNL | |
| GAB1 | Q13480 | Y627 | KGDKQVEYLDLDLDS | |
| GAB1 | Q13480 | Y659 | VADERVDYVVVDQQK | |
| Met | P08581 | Y1003 | VSNESVDYRATFPED | + |
| Met | P08581 | Y1230 | FGLARDMYDKEYYSV | + |
| Met | P08581 | Y1234 | RDMYDKEYYSVHNKT | + |
| Met | P08581 | Y1235 | DMYDKEYYSVHNKTG | + |
| Met | P08581 | Y1349 | STFIGEHYVHVNATY | - |
| Met | P08581 | Y1356 | YVHVNATYVNVKCVA | - |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
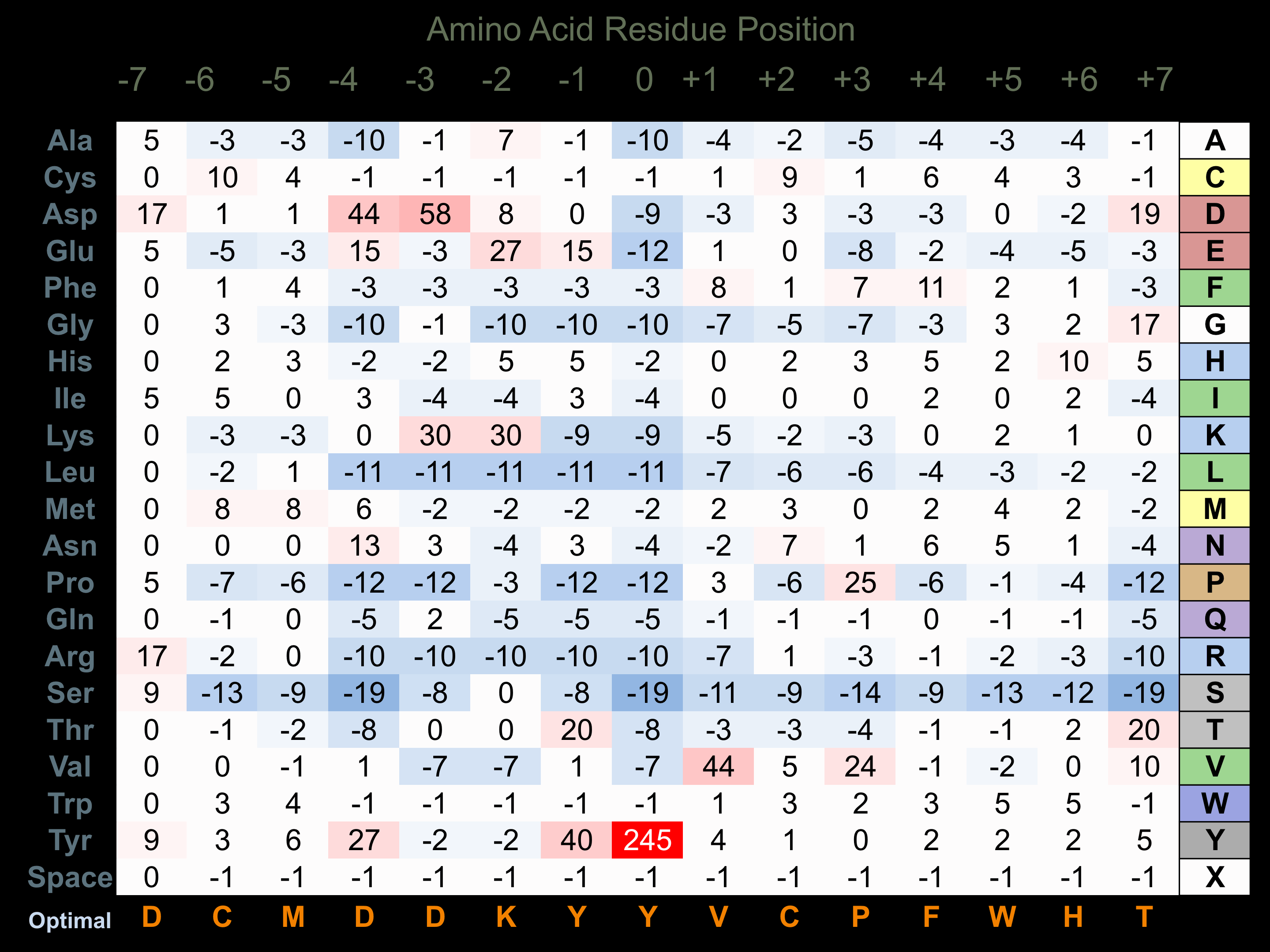
Matrix Type:
Experimentally derived from alignment of 17 known protein substrate phosphosites and 100 peptides phosphorylated by recombinant Met in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, neurological disorders
Specific Diseases (Non-cancerous):
Hypermethioninemia (MET)
Comments:
Hypermethioninemia (MET) arises from abnormal amounts of methionine, an amino acid, in the blood. MET can be affiliated with the liver, brain (overall), and cortex of the brain.
Specific Cancer Types:
Hepatocellular carcinomas (HCC); Renal cell carcinomas, papillary, 1, Familial and somatic; Papillary renal carcinomas; Hepatocellular carcinomas, somatic; Renal cell carcinomas; Papillary carcinomas; Malignant mesothelioma; lung cancer; Salivary gland cancer; Hepatocellular carcinomas, Childhood; Papillary renal cell carcinomas (RCCP); Hepatoblastoma, somatic; Colorectal cancer; Osteosarcoma; Cholangiocarcinomas (CCA); Chordoma; Spinal Chordoma; Alveolar Soft Part Sarcoma (ASPS); Inflammatory Breast carcinomas (IBC); Inflammatory Breast carcinomas; Acral lentiginous melanomas (ALM); Gastric cardia adenocarcinomas; Tall cell variant papillary carcinomas
Comments:
MET is a known oncoprotein (OP). Cancer-related mutations in human tumours point to a gain of function of the protein kinase. The active form of the protein kinase normally acts to promote tumour cell proliferation. Hepatocellular carcinoma (HCC) occurs with higher risk in those that have chronic HCV (hepatitis C virus) or HBV (hepatitis B virus) infection, or aflatoxin exposure in diet, or alcohol-induced cirrhosis, or another form of cirrhosis. HCC is a tumour of the epithelial liver cells. Renal Cell Carcinoma is a kidney carcinoma arising from the proximal renal tube. Malignant Mesothelioma is a carcinoma of the mesothelial tissue (a membrane encapsulating various body cavities like the peritoneum around the abdomen) which can affect breast, lung, and testis tissues. Salivary Gland Cancer is a rare head and neck cancer which may be asymptomatic. Lumps can occur in the ear, cheek, jaw, or lip. Papillary Renal Cell Carcinoma (RCCP) is a rare kidney cancer and can affect the kidney, thyroid, or brain. Osteosarcoma accounts for the most bone marrow cancers in youth, but can also be associated with lung and breast tissue. The rare liver cancer Cholangiocarcinoma (CCA) arises from transformed epithelial cells and may include symptoms such as abnormally dark urine, and itching alongside tumour development. Chordoma is a cancer which will develop from the notochord, which is essential for spine development, but typically will not be diagnosed until later in life. Chordoma is characterized by invading surrounding bone, not metastasizing throughout the body, and leading to headaches, and double vision. Alveolar Soft Part Sarcoma (ASPS) is related to the sarcoma and granular cell tumour disorders. ASPS is a soft-tissue cancer of unknown origin and which can affect the tongue, cervix, or breast. A rare breast cancer, Inflammatory Breast Carcinoma (IBC), is characterized by the formation of a blockage in the lymph nodes, leading to generally inflamed tissue. The rare skin melanoma Acral Lentiginous Melanoma (ALM) can arise from melanocytes, typically on the soles of the feet, palms of the hands, under a nail, or in oral mucosa. ALM does not appear to have a link to sunlight for causing cancer.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +98, p<(0.0003); Brain oligodendrogliomas (%CFC= +50, p<0.044); Breast epithelial carcinomas (%CFC= -62, p<0.004); Clear cell renal cell carcinomas (cRCC) (%CFC= +95, p<0.091); Colon mucosal cell adenomas (%CFC= +234, p<0.0001); Malignant pleural mesotheliomas (MPM) tumours (%CFC= +143, p<0.001); Oral squamous cell carcinomas (OSCC) (%CFC= +51, p<0.0006); Ovary adenocarcinomas (%CFC= -78, p<0.003); Papillary thyroid carcinomas (PTC) (%CFC= +192, p<0.051); Pituitary adenomas (ACTH-secreting) (%CFC= -53, p<0.08); Prostate cancer - primary (%CFC= +93, p<0.0001); Skin melanomas - malignant (%CFC= +120, p<0.0001); and Uterine leiomyomas from fibroids (%CFC= -47, p<0.002). The COSMIC website notes an up-regulated expression score for MET in diverse human cancers of 515, which is 1.1-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 4 for this protein kinase in human cancers was 0.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.11 % in 36569 diverse cancer specimens. This rate is a modest 1.48-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.42 % in 1611 large intestine cancers tested; 0.26 % in 1678 upper aerodigestive tract cancers tested; 0.24 % in 1749 skin cancers tested; 0.22 % in 1744 kidney cancers tested; 0.19 % in 749 endometrium cancers tested; 0.17 % in 565 urinary tract cancers tested; 0.14 % in 4365 lung cancers tested; 0.13 % in 954 stomach cancers tested; 0.11 % in 1481 liver cancers tested; 0.09 % in 1332 ovary cancers tested; 0.05 % in 3043 haematopoietic and lymphoid cancers tested; 0.05 % in 2134 central nervous system cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: Y1253D (47); T1010I (31); M1268T (11); Y1248C (9); R988C (6).
Comments:
In human tumours, the muations are particularly prevalent in 3 regions: amino acid residues 369-375; 982-1010; and 1246-1270.