Nomenclature
Short Name:
CDK4
Full Name:
Cyclin-dependent kinase 4
Alias:
- CRK3
- Cyclin-dependent kinase 4
- EC 2.7.11.22
- Kinase Cdk4
- PSK-J3
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
CDK
SubFamily:
CDK4
Specific Links
Structure
Mol. Mass (Da):
33,730
# Amino Acids:
303
# mRNA Isoforms:
2
mRNA Isoforms:
33,730 Da (303 AA; P11802); 20,725 Da (183 AA; P11802-2)
4D Structure:
Forms a stable complex with D-type G1 cyclins. Interacts with CCND1, SEI1 and ZNF655/VIK.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 6 | 295 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
A2.
Threonine phosphorylated:
T172+, T277.
Tyrosine phosphorylated:
Y17-.
Ubiquitinated:
K22, K35, K106, K118, K142, K155.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 65
65
1133
18
872
 21
21
375
10
157
 13
13
225
8
95
 24
24
421
50
464
 61
61
1069
14
887
 24
24
426
46
274
 27
27
474
19
627
 58
58
1016
27
1561
 41
41
721
10
536
 17
17
297
45
101
 14
14
248
21
140
 57
57
1003
123
899
 14
14
241
19
76
 16
16
283
9
95
 15
15
268
18
137
 25
25
432
8
123
 42
42
738
109
4005
 14
14
239
15
94
 14
14
253
50
127
 49
49
860
56
734
 12
12
214
17
121
 15
15
268
19
128
 14
14
246
17
124
 8
8
139
15
87
 12
12
211
17
76
 55
55
972
35
997
 15
15
261
22
72
 13
13
234
15
103
 23
23
398
15
177
 9
9
156
14
83
 68
68
1202
18
735
 35
35
607
21
713
 100
100
1755
57
2012
 59
59
1033
31
753
 8
8
134
22
97
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
100 99.6
99.6
99.6
100 -
-
-
97 -
-
-
98 97
97
97.6
97 -
-
-
- 94.7
94.7
97
95 95
95
97.3
95 -
-
-
- -
-
-
- 65.3
65.3
75.7
- 72
72
79.9
79 71.9
71.9
80.1
74 -
-
-
- -
-
-
- 35.2
35.2
50.3
- -
-
-
- 48.7
48.7
60.8
- -
-
-
- 43.8
43.8
62.7
- -
-
-
- 45.5
45.5
64
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
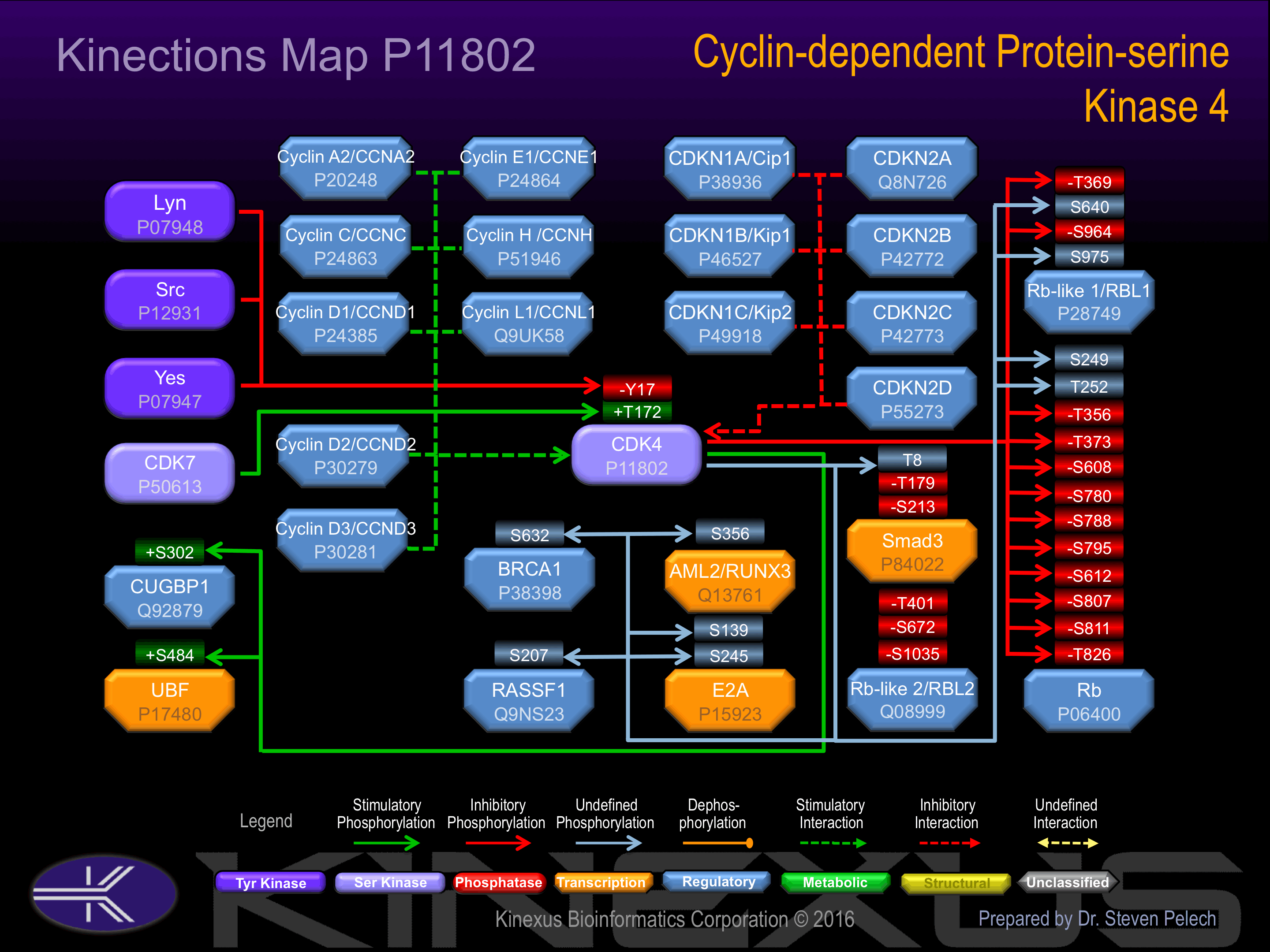
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CDC7 - O00311 |
| 2 | MCM2 - P49736 |
| 3 | CCND1 - P24385 |
| 4 | CDKN2A - P42771 |
| 5 | CDKN2A - Q8N726 |
| 6 | CDC45L - O75419 |
| 7 | CCND3 - P30281 |
| 8 | ORC3L - Q9UBD5 |
| 9 | CDKN1B - P46527 |
| 10 | CCND2 - P30279 |
| 11 | CDC37 - Q16543 |
| 12 | MYOD1 - P15172 |
| 13 | SERTAD1 - Q9UHV2 |
| 14 | PCNA - P12004 |
| 15 | CDKN2B - P42772 |
Regulation
Activation:
Phosphorylation of Thr-172 increases phosphotransferase activity.
Inhibition:
Phosphorylation of Tyr-17 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| AML2 | Q13761 | S356 | SSSGGDRSPTRMLAS | |
| BRCA1 | P38398 | S632 | LVVSRNLSPPNCTEL | |
| CUGBP1 | Q92879 | S302 | TSSGSSPSSSSSNSV | + |
| E2A | P15923 | S139 | LNSPGPLSPSGMKGT | |
| E2A | P15923 | S245 | PMLGGGSSPLPLPPG | |
| FOXM1 | Q08050 | T611 | ETLPISSTPSKSVLP | + |
| RASSF1 | Q9NS23 | S207 | TSVRRRTSFYLPKDA | |
| Rb | P06400 | S249 | AVIPINGSPRTPRRG | |
| Rb | P06400 | S608 | TAADMYLSPVRSPKK | - |
| Rb | P06400 | S780 | STRPPTLSPIPHIPR | - |
| Rb | P06400 | S788 | PIPHIPRSPYKFPSS | - |
| Rb | P06400 | S795 | SPYKFPSSPLRIPGG | - |
| Rb | P06400 | S807 | PGGNIYISPLKSPYK | - |
| Rb | P06400 | S811 | IYISPLKSPYKISEG | - |
| Rb | P06400 | T252 | PINGSPRTPRRGQNR | |
| Rb | P06400 | T356 | DSFETQRTPRKSNLD | - |
| Rb | P06400 | T373 | VNVIPPHTPVRTVMN | - |
| Rb | P06400 | T826 | LPTPTKMTPRSRILV | - |
| Rb-like 1 | P28749 | S640 | RRDMQPLSPISVHER | |
| Rb-like 1 | P28749 | S964 | MMDAPPLSPFPHIKQ | - |
| Rb-like 1 | P28749 | S975 | HIKQQPGSPRRISQQ | |
| Rb-like 1 | P28749 | T369 | KRSFAPSTPLTGRRY | - |
| Rb-like 2 | Q08999 | S1035 | NMDAPPLSPYPFVRT | - |
| Rb-like 2 | Q08999 | S672 | TLYDRYSSPPASTTR | - |
| Rb-like 2 | Q08999 | T401 | SKALRISTPLTGVRY | - |
| Smad3 | P84022 | S213 | NLSPNPMSPAHNNLD | - |
| Smad3 | P84022 | T179 | PQSNIPETPPPGYLS | - |
| Smad3 | P84022 | T8 | MSSILPFTPPIVKRL | |
| UBF | P17480 | S484 | ERGKLPESPKRAEEI | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
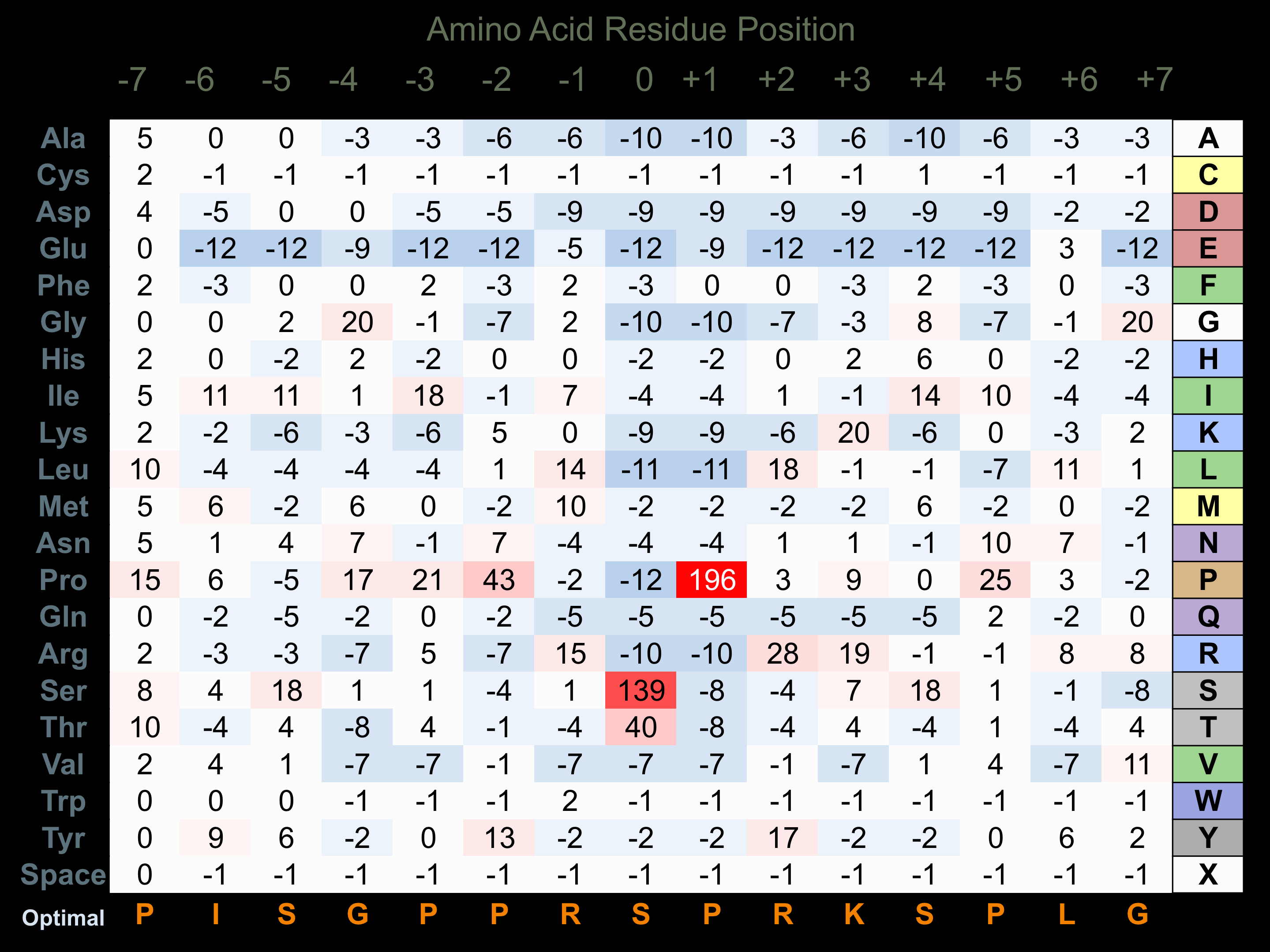
Matrix Type:
Experimentally derived from alignment of 48 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
CDK4-linked melanomas; Astrocytomas; Glioblastomas multiforme; melanomas, cutaneous malignant, 3; Liposarcoma; Familial melanomas; Retinoblastomas; Oligodendrogliomas; Esophageal cancer; Lipoma; Gliosarcomas; Pilocytic astrocytomas; Anaplastic oligodendrogliomas; Dedifferentiated liposarcomas; Osteosarcomas; Atypical lipomatous tumours; Cutaneous malignant melanomas; Atypical mole syndrome; Chondromas; Pleomorphic xanthoastrocytomas; Female breast carcinomas; Embryonal sarcomas; Plasma cell leukemias; Ewing's family of tumours; Spindle cell lipomas; Chordoid gliomas; Retroperitoneal sarcoma; Undifferentiated embryonal sarcoma of the liver; Superficial spreading melanomas; Mixed-type liposarcoma; Spindle cell liposarcomas; Juxtacortical osteosarcomas; Melanomas, malignant, somatic; CDK4-related cutaneous malignant melanomas
Comments:
CDK4 appears to be an oncoprotein (OP). Inactivating germline mutations in the p16 gene have been observed in ~50% of patients with familial hereditary melanoma, indicating a potential role for elevated CDK4 activity in tumorigenesis. Mutations in the p16 gene have been observed in small cell lung carcinomas as well as other cancer types. In addition, several mutations in the CDK4 gene seqeunce have been identified in melanoma patients, including a somatic R24C substitution and a germline R24H substitution. The somatic R24C mutation prevents p16 binding and inhibition of the CDK4 protein, but does not affect the CDK4-cyclin-D interation or the kinase catalytic activity of the protein. Both the R24C and inactivating p16 mutations are predicted to result in the oncogenic activation of the CDK4 protein, as the p16-mediated negative regualtion has been eliminated. In addition to melanoma, CDK4 expression is also required for the tumorigenesis of primary epithelial cells. In the resulting epithelial tumour cells, CDK4 activity appears to be stimulated by the oncogenic activity of RAS, highlighting the potential importance of CDK4 suppression for the prevention of uncontrolled epithelial cell growth. In addition, CDK4 activity is necessary for RAS-mediated transformation in a variety of cancer types. Furthermore, over-expression of both the CDK4 gene and protein are observed in a significant proportion of human breast cancer specimens, corresponding with elevated levels of the cyclin D1 protein. Cyclin D1 has been shown to have a critical role in ErbB2- and Ras-mediated breast cancer tumorigenesis, indicating that an interaction between CDK4 and cyclin D1 might promote breast cancer development. Additionally, elevated expression of CDK4 is also commonly observed in sarcoma and glioma cancer cells. In general, CDK4 levels are up-regulated 1.6-fold in human tumours compared to most other protein kinases. Therefore, the CDK4 protein appears to have a key role in the promotion of tumorigenesis, either due to a mutation that prevents p16-mediated inhibition of the CDK4 protein or resulting from the elevated expression of CDK4 that effectively overwhelms the inhibitory capacity of the normal levels of p16 protein.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +94, p<0.0001); Cervical cancer (%CFC= -64, p<0.0001); Classical Hodgkin lymphomas (%CFC= +165, p<0.003); Colon mucosal cell adenomas (%CFC= +126, p<0.0001); Gastric cancer (%CFC= +80, p<0.0002); Large B-cell lymphomas (%CFC= +275, p<0.001); Lung adenocarcinomas (%CFC= +53, p<0.006); Malignant pleural mesotheliomas (MPM) tumours (%CFC= +133, p<0.0001); Oral squamous cell carcinomas (OSCC) (%CFC= +173, p<0.022); Skin fibrosarcomas (%CFC= +134); Skin melanomas - malignant (%CFC= +177, p<0.0001); and Vulvar intraepithelial neoplasia (%CFC= +47, p<0.012); The COSMIC website notes an up-regulated expression score for CDK4 in diverse human cancers of 730, which is 1.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 24 for this protein kinase in human cancers was 0.4-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 26351 diverse cancer specimens. This rate is only -11 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.3 % in 1335 large intestine cancers tested.
Frequency of Mutated Sites:
None > 4 in 21,410 cancer specimens
Comments:
No deletions, insertions or complex mutations are noted on the COSMIC website.