Nomenclature
Short Name:
STLK3
Full Name:
STE20-SPS1-related proline-alanine rich protein kinase
Alias:
- DCHT
- EC 2.7.11.1
- STE20/SPS1-related proline-alanine rich protein kinase
- STK39
- Serine threonine kinase 39
- Serine/threonine-protein kinase 39
- SPAK
- Ste-20 related kinase
Classification
Type:
Protein-serine/threonine kinase
Group:
STE
Family:
STE20
SubFamily:
FRAY
Structure
Mol. Mass (Da):
59,642
# Amino Acids:
547
# mRNA Isoforms:
2
mRNA Isoforms:
59,474 Da (545 AA; Q9UEW8); 57,937 Da (526 AA; Q9UEW8-2)
4D Structure:
NA
1D Structure:
Subfamily Alignment
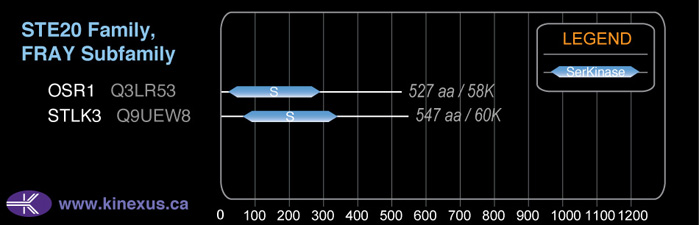
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 63 | 337 | Pkinase |
| 362 | 368 | Nuclear Localization |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
A2, K349.
Serine phosphorylated:
S309+, S311+, S323+, S325+, S370, S371+, S373+, S385, S387, S430, S448.
Threonine phosphorylated:
T219, T224, T231+, T233+, T235+, T237-, T354.
Tyrosine phosphorylated:
Y63, Y273, Y275, Y276, Y306, Y444.
Ubiquitinated:
K83, K98, K349.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 16
16
838
16
1003
 1
1
49
10
20
 40
40
2055
18
1247
 37
37
1912
69
2482
 16
16
797
14
678
 2
2
81
37
47
 13
13
689
23
552
 36
36
1861
44
2682
 16
16
816
10
645
 5
5
246
73
394
 6
6
320
33
420
 11
11
576
133
638
 20
20
1006
29
1037
 2
2
86
9
47
 23
23
1176
31
1679
 2
2
120
8
111
 7
7
351
202
997
 100
100
5131
25
4849
 3
3
141
66
220
 16
16
803
56
766
 73
73
3762
29
3781
 17
17
852
31
968
 48
48
2467
19
1798
 76
76
3911
25
3638
 29
29
1495
29
1601
 30
30
1527
49
1970
 34
34
1720
32
1951
 73
73
3728
26
3699
 16
16
824
25
760
 0.5
0.5
28
14
25
 25
25
1276
18
765
 49
49
2494
21
4430
 0.3
0.3
16
57
89
 24
24
1220
26
960
 26
26
1344
22
1173
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 74.2
74.2
75
100 27.4
27.4
45.2
96 -
-
-
- -
-
-
- 73.6
73.6
76.6
98 -
-
-
- 96
96
96.4
96.5 96.4
96.4
96.9
96.5 -
-
-
- 89.8
89.8
92.3
- 79.9
79.9
81.5
95 28
28
46.3
87 28.3
28.3
45.3
78 -
-
-
- 56.9
56.9
71.7
- 47.4
47.4
59.1
- -
-
-
- 54.3
54.3
71.7
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | WNK4 - Q96J92 |
| 2 | MAPK14 - Q16539 |
| 3 | AATK - Q6ZMQ8 |
| 4 | SLC12A2 - P55011 |
| 5 | RELT - Q969Z4 |
| 6 | HSPH1 - Q92598 |
| 7 | PRKCQ - Q04759 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| SLC12A2 (NKCC1) | P55011 | T203 | HQHYYYDTHTNTYYL | + |
| SLC12A2 (NKCC1) | P55011 | T207 | YYDTHTNTYYLRTFG | + |
| SLC12A2 (NKCC1) | P55011 | T212 | TNTYYLRTFGHNTMD | + |
| SLC12A2 (NKCC1) | P55011 | T217 | LRTFGHNTMDAVPRI | + |
| SLC12A3 | P55017 | T46 | SSHPSHLTHSSTFCM | |
| SLC12A3 | P55017 | T55 | SSTFCMRTFGYNTID | |
| SLC12A3 | P55017 | T60 | MRTFGYNTIDVVPTY | |
| STLK3 (STK39; SPAK) | Q9UEW8 | T233 | TRNKVRKTFVGTPCW | + |
| STLK3 (STK39; SPAK) | Q9UEW8 | T237 | VRKTFVGTPCWMAPE | - |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
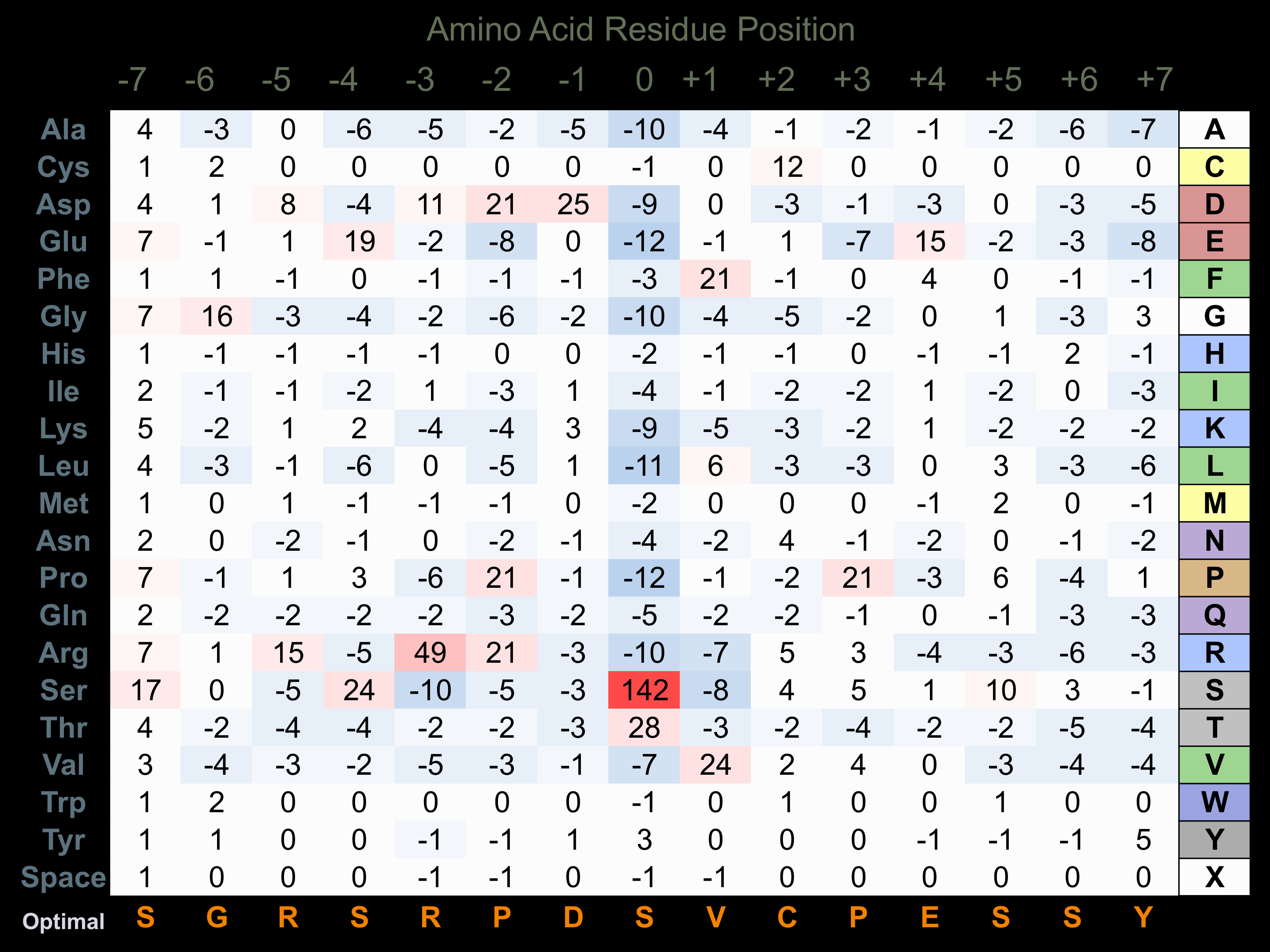
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Staurosporine | IC50 > 50 nM | 5279 | 22037377 | |
| Sunitinib | Kd = 140 nM | 5329102 | 535 | 19654408 |
| NVP-TAE684 | Kd = 150 nM | 16038120 | 509032 | 22037378 |
| SU14813 | Kd = 430 nM | 10138259 | 1721885 | 22037378 |
| Nintedanib | Kd = 520 nM | 9809715 | 502835 | 22037378 |
| Dovitinib | Kd = 530 nM | 57336746 | 22037378 | |
| KW2449 | Kd = 890 nM | 11427553 | 1908397 | 22037378 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| Lestaurtinib | Kd = 1.1 µM | 126565 | 22037378 | |
| AST-487 | Kd = 1.5 µM | 11409972 | 574738 | 22037378 |
| TG101348 | Kd = 1.6 µM | 16722836 | 1287853 | 22037378 |
| Ruxolitinib | Kd = 1.9 µM | 25126798 | 1789941 | 22037378 |
| Bosutinib | Kd = 3.7 µM | 5328940 | 288441 | 22037378 |
Disease Linkage
General Disease Association:
Endocrine disorder
Specific Diseases (Non-cancerous):
Glanders
Comments:
STLK3 is linked to Glanders, which is a bacterial infectious disease with symptoms including nodular lesions in the lung, headaches, and mucopurulent nasal discharge. Glanders can affect lung tissue, eyes, and testes.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +136, p<0.0005); Breast epithelial carcinomas (%CFC= +106, p<0.077); Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= +101, p<0.07); Cervical epithelial cancer (%CFC= +82, p<0.002); Cervical cancer (%CFC= +99, p<0.0005); Large B-cell lymphomas (%CFC= +114, p<0.014); Lung adenocarcinomas (%CFC= +89, p<0.006); Pituitary adenomas (ACTH-secreting) (%CFC= +81); Prostate cancer (%CFC= +63, p<0.038); and Skin melanomas - malignant (%CFC= -71, p<0.004).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24433 diverse cancer specimens. This rate is only -2 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: Y182C (8).
Comments:
Only 2 deletions, and no insertions or complex mutations are noted on the COSMIC website.

