Nomenclature
Short Name:
RSK2
Full Name:
Ribosomal protein S6 kinase alpha 3
Alias:
- 90 kDa ribosomal protein S6 kinase 3
- CLS
- ISPK-1
- KS6A3
- MAPKAPK1B
- MRX19; pp90RSK2; RPS6KA3; S6K-alpha 3
- EC 2.7.11.1
- HU-3
- Insulin-stimulated protein kinase 1
- ISPK1
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
RSK
SubFamily:
RSK
Specific Links
Structure
Mol. Mass (Da):
83,736
# Amino Acids:
740
# mRNA Isoforms:
1
mRNA Isoforms:
83,736 Da (740 AA; P51812)
4D Structure:
Forms a complex with either ERK1 or ERK2 in quiescent cells. Transiently dissociates following mitogenic stimulation By similarity. Interacts with NFATC4
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
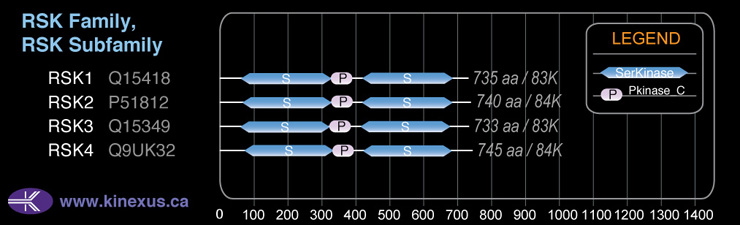
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K81, K100, K103.
Serine phosphorylated:
S17, S19, S78-, S227+, S325, S328, S369+, S375, S386+, S392, S396, S415, S517, S556, S708, S715, S737.
Threonine phosphorylated:
T231-, T278, T362, T365, T391, T478, T493, T577+, T581-, T625, T706.
Tyrosine phosphorylated:
Y144, Y226+, Y234-, Y356, Y433, Y470, Y483, Y488, Y529, Y547, Y580+, Y644, Y688, Y707.
Ubiquitinated:
K72, K322, K504, K658.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 26
26
1083
25
1057
 2
2
63
13
37
 3
3
125
1
0
 26
26
1063
93
2192
 21
21
887
24
632
 5
5
204
58
249
 14
14
594
33
576
 58
58
2400
44
3281
 16
16
670
10
643
 4
4
173
89
148
 3
3
132
24
189
 14
14
592
129
647
 3
3
127
12
23
 2
2
68
12
37
 4
4
165
21
258
 3
3
112
14
35
 4
4
156
381
892
 2
2
76
10
46
 10
10
434
67
270
 23
23
957
84
768
 2
2
83
20
114
 5
5
195
22
381
 3
3
118
10
43
 0.9
0.9
37
10
27
 7
7
305
20
626
 48
48
2007
62
2641
 2
2
96
15
30
 2
2
68
10
49
 2
2
72
10
55
 2
2
81
28
60
 25
25
1042
18
553
 100
100
4167
26
7174
 5
5
213
105
313
 27
27
1131
57
777
 2
2
103
35
121
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
96 76.7
76.7
86.3
98 -
-
-
100 -
-
-
- 94.5
94.5
95.2
100 -
-
-
- 99.8
99.8
99.8
100 80.8
80.8
88.6
100 -
-
-
- 93.2
93.2
94.1
- 80.1
80.1
87.5
97 77.4
77.4
86
- 82
82
89.3
86 -
-
-
- 24.3
24.3
37.2
63 -
-
-
- 57.6
57.6
71.5
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | MAPK1 - P28482 |
| 2 | MAPK3 - P27361 |
| 3 | FGF2 - P09038 |
| 4 | BAD - Q92934 |
| 5 | MAPK14 - Q16539 |
| 6 | CREB1 - P16220 |
| 7 | SHANK1 - Q9Y566 |
| 8 | METTL1 - Q9UBP6 |
| 9 | ESR1 - P03372 |
| 10 | STK11 - Q15831 |
| 11 | GRIN2D - O15399 |
Regulation
Activation:
Phosphorylation at Ser-386 permits recruitment and activation of PDK1 by autophosphorylation to then phosphorylate S227. Phosphorylation at Ser-227, Ser-369 and Thr-577 increases phosphotransferase activity. Phosphorylation at Tyr-529 induces interaction with the upstream kinases ERK1and ERK2, as well as FGFR3.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| RSK2 | P51812 | S227 | DHEKKAYSFCGTVEY | + |
| PDK1 | O15530 | S227 | DHEKKAYSFCGTVEY | + |
| PDK1 | O15530 | S369 | TAKTPKDSPGIPPSA | + |
| RSK2 | P51812 | S386 | HQLFRGFSFVAITSD | + |
| ERK1 | P27361 | S386 | HQLFRGFSFVAITSD | + |
| PDK1 | O15530 | S386 | HQLFRGFSFVAITSD | + |
| p70S6K | P23443 | S386 | HQLFRGFSFVAITSD | + |
| FYN | P06241 | Y470 | EIEILLRYGQHPNII | |
| SRC | P12931 | Y470 | EIEILLRYGQHPNII | |
| SRC | P12931 | Y483 | IITLKDVYDDGKYVY | |
| FYN | P06241 | Y483 | IITLKDVYDDGKYVY | |
| SRC | P12931 | Y488 | DVYDDGKYVYVVTEL | |
| FGFR3 | P22607 | Y488 | DVYDDGKYVYVVTEL | |
| FYN | P06241 | Y488 | DVYDDGKYVYVVTEL | |
| FGFR3 | P22607 | Y529 | TITKTVEYLHAQGVV | ? |
| FYN | P06241 | Y529 | TITKTVEYLHAQGVV | ? |
| SRC | P12931 | Y529 | TITKTVEYLHAQGVV | ? |
| ERK2 | P28482 | T577 | AENGLLMTPCYTANF | + |
| ERK1 | P27361 | T577 | AENGLLMTPCYTANF | + |
| PDK1 | O15530 | T577 | AENGLLMTPCYTANF | + |
| FYN | P06241 | Y580 | GLLMTPCYTANFVAP | + |
| SRC | P12931 | Y580 | GLLMTPCYTANFVAP | + |
| FYN | P06241 | Y644 | KFSLSGGYWNSVSDT | |
| SRC | P12931 | Y644 | KFSLSGGYWNSVSDT | |
| FYN | P06241 | Y707 | KGAMAATYSALNRNQ | |
| SRC | P12931 | Y707 | KGAMAATYSALNRNQ |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| 5-HT(2A) | P28223 | S314 | TGRRTMQSISNEQKA | |
| ATF4 | P18848 | S245 | TRGSPNRSLPSPGVL | |
| Bad | Q92934 | S75 | EIRSRHSSYPAGTED | - |
| Bad | Q92934 | S99 | PFRGRSRSAPPNLWA | - |
| CREB1 | P16220 | S133 | EILSRRPSYRKILND | + |
| DAPK1 | P53355 | S289 | QALSRKASAVNMEKF | - |
| eEF2K | O00418 | S366 | SPRVRTLSGSRPPLL | - |
| eIF2A | P05198 | S52 | MILLSELSRRRIRSI | - |
| ERa (ESR1) | P03372 | S167 | GGRERLASTNDKGSM | ? |
| Fos | P01100 | S362 | AAAHRKGSSSNEPSS | + |
| Fos | P01100 | S374 | PSSDSLSSPTLLAL_ | + |
| GSK3a | P49840 | S21 | SGRARTSSFAEPGGG | - |
| GSK3b | P49841 | S9 | SGRPRTTSFAESCKP | - |
| H3.2 | P84228 | S11 | TKQTARKSTGGKAPR | |
| H3.3 | P84243 | S28 | ATKAARKSAPSTGGV | |
| HMGN1 (HMG14) | P05114 | S6 | __PKRKVSSAEGAAK | |
| HSP27 | P04792 | S78 | PAYSRALSRQLSSGV | + |
| HSP27 | P04792 | S82 | RALSRQLSSGVSEIR | ? |
| MEF2C | Q06413 | S183 | SLQRNSMSPGVTHRP | |
| METTL1 | Q9UBP6 | S27 | YYRQRAHSNPMADHT | - |
| NFAT3 | Q14934 | S281 | SGTPSSASPALSRRG | |
| NFAT3 | Q14934 | S285 | SSASPALSRRGSLGE | |
| NFAT3 | Q14934 | S289 | PALSRRGSLGEEGSE | |
| NFAT3 | Q14934 | S344 | QAVALPRSEEPASCN | |
| NFAT3 | Q14934 | S676 | SNGRRKRSPTQSFRF | + |
| nNOS | P29475 | S847 | SYKVRFNSVSSYSDS | |
| Nur77 | P22736 | S351 | GRRGRLPSKPKQPPD | - |
| p27Kip1 | P46527 | S10 | NVRVSNGSPSLERMD | - |
| p27Kip1 | P46527 | T198 | PGLRRRQT_______ | - |
| PFKFB2 | O60825 | S466 | PVRMRRNSFTPLSSS | |
| PFKFB2 | O60825 | S483 | IRRPRNYSVGSRPLK | |
| PKR (PRKR; EIF2AK2) | P19525 | T451 | KRTRSKGTLRYMSPE | - |
| PLD1 | Q13393 | T147 | PIPTRRHTFRRQNVR | |
| PPP1R3A | Q16821 | S46 | PQPSRRGSDSSEDIY | |
| PPP1R3A | Q16821 | S65 | SSGTRRVSFADSFGF | |
| RANBP3 | Q9H6Z4 | S126 | VKRERTSSLTQFPPS | |
| RPS6 | P62753 | S235 | IAKRRRLSSLRASTS | + |
| RPS6 | P62753 | S236 | AKRRRLSSLRASTSK | + |
| RSK2 (RPS6KA3) | P51812 | S227 | DHEKKAYSFCGTVEY | + |
| RSK2 (RPS6KA3) | P51812 | S386 | HQLFRGFSFVAITSD | + |
| STAT3 | P40763 | S727 | NTIDLPMSPRTLDSL | - |
| TH | P07101 | S71 | RFIGRRQSLIEDARK | + |
| TIF-IA | Q9NYV6 | S649 | PVLYMQPSPL_____ |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
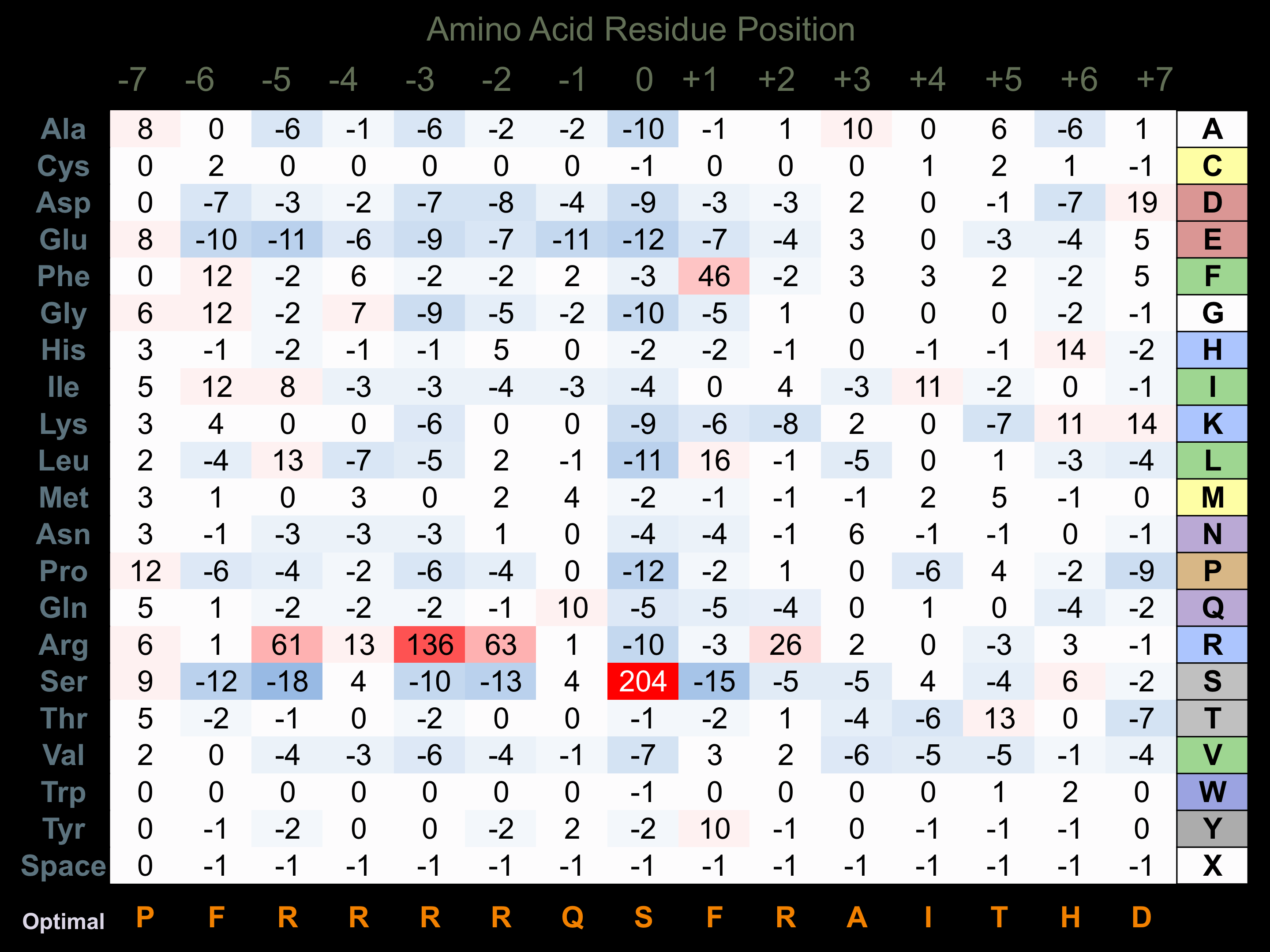
Matrix Type:
Experimentally derived from alignment of 52 known protein substrate phosphosites and 19 peptides phosphorylated by recombinant RSK2 in vitro tested in-house by Kinexus.
Domain #:
2
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Genetic disorders
Specific Diseases (Non-cancerous):
Coffin-Lowry syndrome; Alpha-thalassemia/mental retardation syndrome; Partington syndrome; Estrogen resistance; Mental retardation, X-linked 17/31, Microduplication
Comments:
Coffin-Lowry Syndrome (CLS) is a rare genetic disorder characterized by craniofacial (head and face) and skeletal abnormalities, mental retardation, short statue, and hypotonia. Characteristic symptoms include hypertonia/spasticity, lens ossification, retinal pigment changes, feeding and respiratory problems, developmental delay, mental retardation, and heart and kidney defects. Several mutations in the RPS6KA3 gene have been observed in patients with CLS. In 86 unrelated families with CLS, 71 distinct disease-associated mutations were identified in the RPS6KA3 gene. The observed mutations included missense mutations (38%), nonsense mutations (20%), splicing errors (18%), and short deletions or insertions (21%). The mutations were distributed throughout the gene and did not display clustering of phenotypic association, indicating a loss-of-function effect on the RPS6KA3 protein. In animal studies, knockout of RPS6KA3 in mice lead to failure of osteoblast differentiation, resulting in abnoramlly delayed bone formation during development resulting in significantly reduced bone mass in the adult mice. Therefore, loss-of-function in the RPS6KA3 protein may play a role in the pathogenesis of Coffin-Lowry syndrome.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +57, p<0.013); and Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +382, p<0.0008). The COSMIC website notes an up-regulated expression score for RSK2 in diverse human cancers of 466, which is very close to the average score of 462 for the human protein kinases. The down-regulated expression score of 125 for this protein kinase in human cancers was 2.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 25463 diverse cancer specimens. This rate is only 23 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.46 % in 1119 large intestine cancers tested; 0.43 % in 1278 liver cancers tested; 0.36 % in 602 endometrium cancers tested; 0.09 % in 1503 breast cancers tested; 0.08 % in 1941 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: Y483C (4); L381P (3).
Comments:
Ten deletions, 4 insertions and 5 complex mutations are noted on the COSMIC website.

