Nomenclature
Short Name:
SGK
Full Name:
Serine-threonine-protein kinase Sgk1
Alias:
- EC 2.7.11.1
- Sgk1
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
SGK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
48956
# Amino Acids:
431
# mRNA Isoforms:
5
mRNA Isoforms:
59,904 Da (526 AA; O00141-2); 52,119 Da (459 AA; O00141-5); 50,623 Da (445 AA; O00141-3); 48,942 Da (431 AA; O00141) and 47,910 Da (421 AA; O00141-4)
4D Structure:
Interacts with NEDD4 and NEDD4L.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
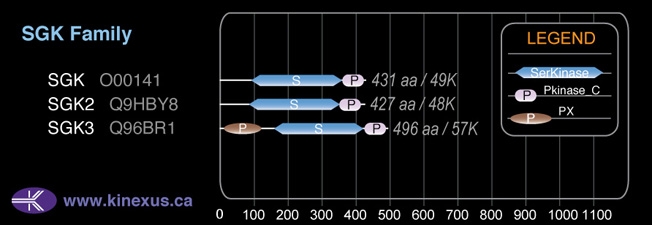
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K41.
Serine phosphorylated:
S74, S78, S377, S397, S401, S422+.
Threonine phosphorylated:
T256+, T369+.
Tyrosine phosphorylated:
Y124.
Ubiquitinated:
K127.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 36
36
1649
16
1587
 21
21
985
10
788
 5
5
236
12
112
 28
28
1307
68
1477
 30
30
1368
14
890
 15
15
708
58
531
 18
18
820
19
530
 24
24
1103
39
1858
 22
22
994
10
711
 20
20
922
56
569
 6
6
264
25
382
 36
36
1660
106
1817
 8
8
391
23
242
 8
8
391
9
272
 7
7
312
11
294
 44
44
2042
8
1949
 11
11
506
114
3614
 4
4
163
19
126
 10
10
457
54
313
 27
27
1226
56
833
 20
20
912
21
826
 3
3
134
22
88
 12
12
537
21
635
 14
14
629
19
489
 3
3
157
21
159
 27
27
1259
48
1433
 11
11
516
26
276
 11
11
514
19
425
 5
5
236
19
186
 0.5
0.5
24
14
21
 21
21
960
18
676
 100
100
4616
21
9681
 29
29
1356
56
1403
 45
45
2082
31
911
 22
22
1023
22
1023
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 69.8
69.8
70.3
96 66.3
66.3
67.2
100 -
-
-
97 -
-
-
98 97.7
97.7
99.1
96 -
-
-
- 96.5
96.5
98.6
94.5 97
97
98.6
97.5 -
-
-
- 95.4
95.4
98.4
- 95.1
95.1
97.2
97 88.9
88.9
94.2
91 86.6
86.6
92.8
89 -
-
-
- 34.7
34.7
48
- -
-
-
- 47.1
47.1
63.5
54 62.7
62.7
75
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 29.1
29.1
41.9
52 -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | SLC9A3R2 - Q15599 |
| 2 | NEDD4L - Q96PU5 |
| 3 | SLC1A3 - P43003 |
| 4 | KCNJ1 - P48048 |
| 5 | CREB1 - P16220 |
| 6 | MAPK7 - Q13164 |
| 7 | CAMKK2 - Q96RR4 |
| 8 | PIK3CA - P42336 |
| 9 | PPP2CA - P67775 |
| 10 | CARHSP1 - Q9Y2V2 |
| 11 | GSK3B - P49841 |
| 12 | GSK3A - P49840 |
| 13 | BRAF - P15056 |
| 14 | PDK2 - Q15119 |
| 15 | ATP5D - P30049 |
Regulation
Activation:
Activated by phosphorylation at Thr-256 and Ser-422.
Inhibition:
NA
Synthesis:
Isoform 1 is induced by serum and/or glucocorticoids.
Degradation:
Isoform 2 not induced by glucocorticoids. By excessive extracellular glucose and by TGF-beta, in cultured cells.
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| B-Raf | P15056 | S365 | GQRDRSSSAPNVHIN | - |
| CaRHSP1 | Q9Y2V2 | S52 | TRRTRTFSATVRASQ | |
| CREB1 | P16220 | S133 | EILSRRPSYRKILND | + |
| Elk-1 | P19419 | S160 | YMRSGLYSTFTIQSL | |
| Elk-1 | P19419 | T161 | MRSGLYSTFTIQSLQ | |
| ENaC alpha | P37088 | S594 | RFRSRYWSPGRGGRG | |
| ERK2 (MAPK1) | P28482 | S29 | GPRYTNLSYIGEGAY | + |
| Fe65 | O00213 | S610 | ECRVRFLSFLAVGRD | |
| FOXO1A | Q12778 | S319 | TFRPRTSSNASTISG | + |
| FOXO1A | Q12778 | T24 | LPRPRSCTWPLPRPE | - |
| FOXO3 (FKHRL1) | O43524 | S253 | APRRRAVSMDNSNKY | - |
| FOXO3 (FKHRL1) | O43524 | S315 | DFRSRTNSNASTVSG | - |
| FOXO3 (FKHRL1) | O43524 | T32 | QSRPRSCTWPLQRPE | |
| GLUT4 | P14672 | S274 | LERERPLSLLQLLGS | |
| Huntingtin | P42858 | S421 | GGRSRSGSIVELIAG | |
| IkBa | P25963 | S32 | LLDDRHDSGLDSMKD | |
| IKKb (IKBKINASE) | O14920 | S181 | DQGSLCTSFVGTLQY | + |
| iNOS | P35228 | S739 | VFTMRLKSRQNLQSP | |
| iNOS | P35228 | S909 | ILKPRFYSISSSRDH | |
| MEKK3 (MAP3K3) | Q99759 | S166 | EPRSRHLSVSSQNPG | - |
| MEKK3 (MAP3K3) | Q99759 | S337 | DPRGRLRSADSENAL | - |
| MKK4 (MAP2K4, MEK4) | P45985 | S80 | IERLRTHSIESSGKL | - |
| NDRG1 | Q92597 | S330 | LMRSRTASGSSVTSL | |
| NDRG1 | Q92597 | T328 | TRLMRSRTASGSSVT | |
| NDRG1 | Q92597 | T346 | GTRSRSHTSEGTRSR | |
| NDRG1 | Q92597 | T356 | GTRSRSHTSEGTRSR | |
| NDRG1 | Q92597 | T366 | GTRSRSHTSEGAHLD | |
| NDRG2 | Q9UN36 | S332 | LSRSRTASLTSAASV | |
| NDRG2 | Q9UN36 | S346 | VDGNRSRSRTLSQSS | |
| NDRG2 | Q9UN36 | T330 | TRLSRSRTASLTSAA | |
| NDRG2 | Q9UN36 | T348 | GNRSRSRTLSQSSES | |
| NEDD4L | Q96PU5 | S342 | SSRLRSCSVTDAVAE | - |
| NEDD4L | Q96PU5 | S428 | AEDGASGSATNSNNH | - |
| NEDD4L | Q96PU5 | S448 | IRRPRSLSSPTVTLS | ? |
| NEDD4L | Q96PU5 | T367 | AGRARSSTVTGGEEP | |
| NHE3 | P48764 | S663 | TMRKRLESFKSTKLG | |
| p27Kip1 | P46527 | T157 | GIRKRPATDDSSTQN | - |
| p27Kip1 | P46527 | T198 | PGLRRRQT_______ | - |
| PIP5K | Q9Y2I7 | S318 | LSLDRSGSPMVPSYE | |
| ROMK | P48048 | S44 | RQRARLVSKDGRCNI | |
| SLC1A6 | P48664 | T43 | RTRLRLQTMTLEHVL | |
| SRF | P11831 | S103 | RGLKRSLSEMEIGMV | |
| SRF | P11831 | S75 | AGALYSGSEGDSESG | |
| SRF | P11831 | S77 | PTAGALYSGSEGDSE | |
| Tau iso8 | P10636-8 | S214 | GSRSRTPSLPTPPTR | |
| Wnk1 (PRKWNK1) | Q9H4A3 | T60 | EYRRRRHTMDKDSRG | + |
| Wnk4 (PRKWNK4) | Q96J92 | S1190 | SSRQRRLSKGSFPTS | - |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
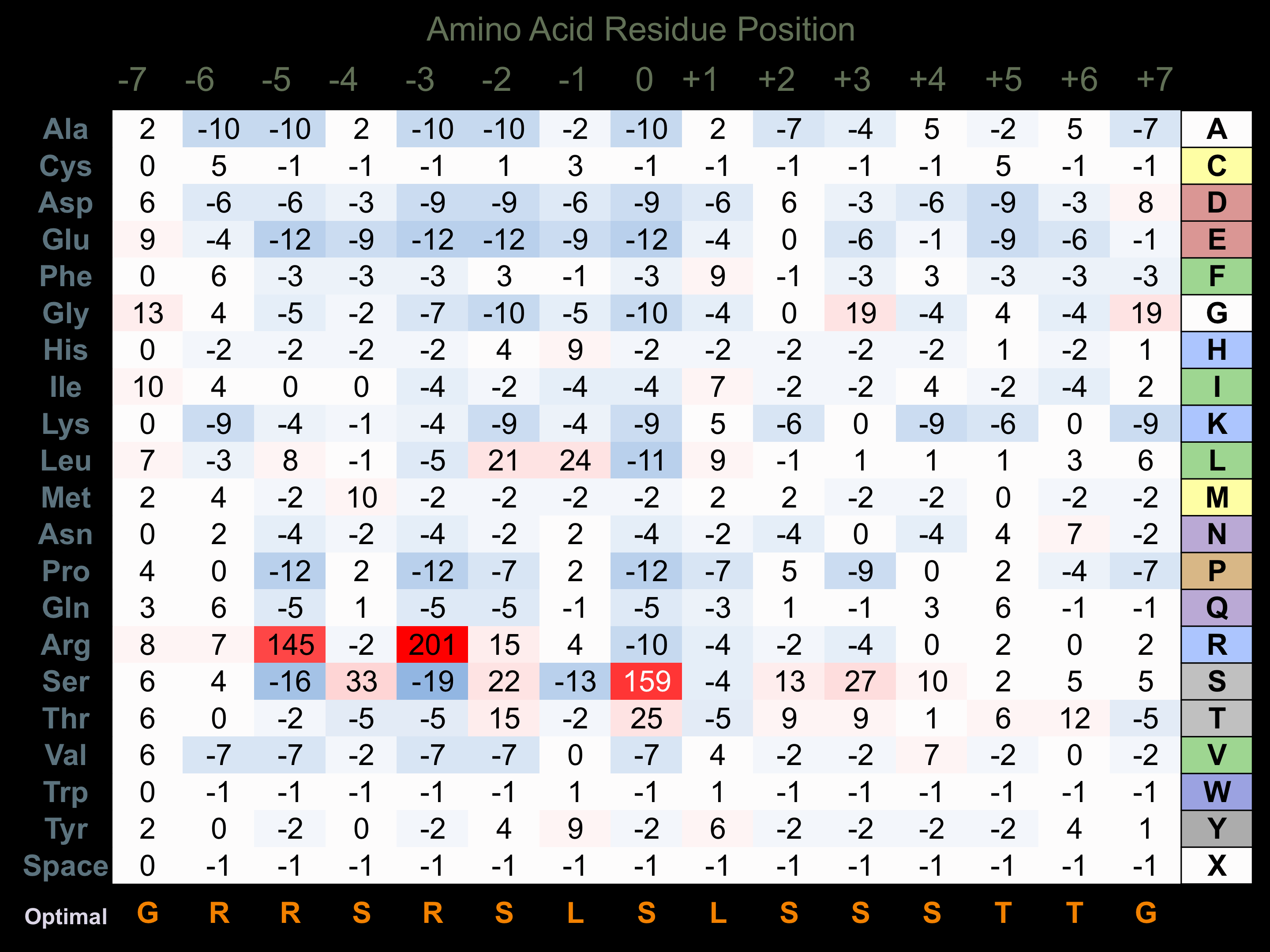
Matrix Type:
Experimentally derived from alignment of 60 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cardiovascular, and nephrological disorders
Specific Diseases (Non-cancerous):
Hypertension (HTN); Diabetic nephropathy
Comments:
Hypertension (HTN) is a rare cardiovascular disorder that manifests as high blood pressure causing a higher risk of stroke, heart failure, heart attack, and kidney failure. SGK kinase phosphotransferase activity can be fully abolished with a K127M mutation, or it can be partly inhibited with one of the T256A, T256D, T256E, or S422A mutations. A 10-fold increase in phosphotransferase activity has been noted with a S422-D mutation. NEDD4 and NEDD4L complex formation with SGK can be inhibited with a Y298A mutation.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= -55, p<0.002); Brain glioblastomas (%CFC= -82, p<0.002); Brain oligodendrogliomas (%CFC= -68, p<0.011); Cervical epithelial cancer (%CFC= +45, p<0.022); Cervical cancer (%CFC= +178, p<0.0001); Classical Hodgkin lymphomas (%CFC= +286, p<(0.0003); Colon mucosal cell adenomas (%CFC= -70, p<0.0001); Colorectal adenocarcinomas (early onset) (%CFC= +72, p<0.058); Gastric cancer (%CFC= -48, p<0.0009); Large B-cell lymphomas (%CFC= +899, p<0.005); Lung adenocarcinomas (%CFC= -47, p<0.0002); Ovary adenocarcinomas (%CFC= -65, p<0.002); Pituitary adenomas (aldosterone-secreting) (%CFC= -54, p<0.057); Skin melanomas - malignant (%CFC= +88, p<0.004); Uterine fibroids (%CFC= -65, p<0.0009); and Uterine leiomyomas from fibroids (%CFC= -79, p<0.083). The COSMIC website notes an up-regulated expression score for SGK in diverse human cancers of 312, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.12 % in 25490 diverse cancer specimens. This rate is a modest 1.54-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.54 % in 603 endometrium cancers tested; 0.31 % in 895 skin cancers tested; 0.3 % in 2096 haematopoietic and lymphoid cancers tested; 0.27 % in 1270 large intestine cancers tested; 0.25 % in 548 urinary tract cancers tested; 0.18 % in 1967 lung cancers tested; 0.18 % in 127 biliary tract cancers tested; 0.16 % in 1276 kidney cancers tested; 0.12 % in 589 stomach cancers tested; 0.12 % in 1512 liver cancers tested; 0.07 % in 710 oesophagus cancers tested; 0.07 % in 1316 breast cancers tested; 0.06 % in 2082 central nervous system cancers tested; 0.05 % in 441 autonomic ganglia cancers tested; 0.04 % in 558 thyroid cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: P77S (3).
Comments:
Ten deletions, 2 insertions and 1 complex mutations are noted on the COSMIC website.

