Nomenclature
Short Name:
p38a
Full Name:
Mitogen-activated protein kinase 14
Alias:
- CRK1
- CSAID binding protein
- Cytokine suppressive anti-inflammatory drug binding protein
- EC 2.7.11.24
- MAPK14
- Kinase p38-alpha; PRKM15; p38a MAPK; SAPK2A
- CSBP
- CSBP1
- CSBP2
- CSPB1
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
MAPK
SubFamily:
p38
Specific Links
Structure
Mol. Mass (Da):
41293
# Amino Acids:
360
# mRNA Isoforms:
5
mRNA Isoforms:
41,493 Da (360 AA; Q16539-2); 41,293 Da (360 AA; Q16539); 35,453 Da (307 AA; Q16539-4); 34,092 Da (297 AA; Q16539-3); 29,388 Da (256 AA; Q16539-5)
4D Structure:
Binds to a kinase interaction motif within the protein tyrosine phosphatase, PTPRR. This interaction retains MAPK14 in the cytoplasm and prevents nuclear accumulation. Interacts with SPAG9 By similarity. Interacts with NP60 and FAM48A.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
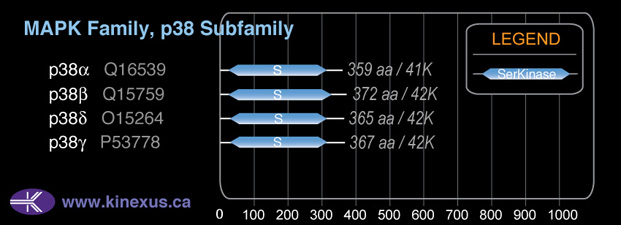
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 24 | 308 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K53, K152.
Serine phosphorylated:
S2, S28, S56, S326.
Threonine phosphorylated:
T7, T16, T123-, T175+, T180+, T185-, T241, T263.
Tyrosine phosphorylated:
Y182+, Y323+.
Ubiquitinated:
K15, K118, K139, K165.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 86
86
1732
55
2934
 4
4
72
32
56
 6
6
115
15
77
 14
14
284
175
430
 34
34
693
48
595
 3
3
62
150
66
 15
15
304
66
507
 23
23
455
72
565
 27
27
554
34
449
 5
5
104
172
117
 5
5
97
59
85
 32
32
640
341
642
 4
4
72
59
47
 3
3
54
26
49
 4
4
78
46
67
 3
3
68
31
46
 3
3
70
319
51
 4
4
77
34
59
 6
6
114
183
82
 30
30
597
218
629
 3
3
69
43
71
 6
6
117
51
112
 3
3
68
35
41
 3
3
56
35
48
 7
7
141
43
126
 27
27
551
103
631
 4
4
85
65
55
 3
3
63
35
54
 5
5
102
35
77
 10
10
203
56
155
 24
24
492
48
350
 100
100
2022
66
4832
 11
11
218
106
250
 44
44
888
114
757
 4
4
78
79
87
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 96.1
96.1
97.8
100 89.4
89.4
90.8
100 -
-
-
100 -
-
-
99 -
-
-
96 -
-
-
- 99.4
99.4
99.4
99 98.6
98.6
99.2
99 -
-
-
- -
-
-
- 96.4
96.4
98.6
92.5 88.4
88.4
94.7
87 85.9
85.9
92.2
89 -
-
-
- 68.5
68.5
80.3
70 75
75
85
- 62.9
62.9
76.7
64 -
-
-
- -
-
-
- -
-
-
- -
-
-
48 -
-
-
46.5 43
43
57.9
54 50.6
50.6
69.4
53
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | MAP2K6 - P52564 |
| 2 | MAPKAPK2 - P49137 |
| 3 | ATF2 - P15336 |
| 4 | DUSP10 - Q9Y6W6 |
| 5 | MAP2K3 - P46734 |
| 6 | MAPK1 - P28482 |
| 7 | MEF2A - Q02078 |
| 8 | DUSP16 - Q9BY84 |
| 9 | DUSP1 - P28562 |
| 10 | CDC25C - P30307 |
| 11 | RPS6KA4 - O75676 |
| 12 | STK39 - Q9UEW8 |
| 13 | CDC25B - P30305 |
| 14 | KRT8 - P05787 |
| 15 | MKNK1 - Q9BUB5 |
Regulation
Activation:
Phosphorylation at Thr-180 and Tyr-182 increases phosphotransferase activity. These phospho-sites are targeted by MAP2K3, MAP2K6, and potentially also MAP2K4. Phosphorylation at Tyr-323 increases phosphotransferase activity.
Inhibition:
Inhibited by dual specificity phosphatases, such as DUSP1. Phosphorylation at Thr-123 inhibits phosphotransferase activity and interaction with MKK6 and MAPKAPK2.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| BARK1 | P25098 | T123 | IVKCQKLTDDHVQFL | - |
| PBK | Q96KB5 | T180 | RHTDDEMTGYVATRW | + |
| MEK6 | P52564 | T180 | RHTDDEMTGYVATRW | + |
| MEK3 | P46734 | T180 | RHTDDEMTGYVATRW | + |
| MEK4 | P45985 | T180 | RHTDDEMTGYVATRW | + |
| p38a | Q16539 | T180 | RHTDDEMTGYVATRW | + |
| PBK | Q96KB5 | Y182 | TDDEMTGYVATRWYR | + |
| MEK6 | P52564 | Y182 | TDDEMTGYVATRWYR | + |
| MEK3 | P46734 | Y182 | TDDEMTGYVATRWYR | + |
| MEK4 | P45985 | Y182 | TDDEMTGYVATRWYR | + |
| p38a | Q16539 | Y182 | TDDEMTGYVATRWYR | + |
| LCK | P06239 | Y323 | DEPVADPYDQSFESR | + |
| FYN | P06241 | Y323 | DEPVADPYDQSFESR | + |
| ZAP70 | P43403 | Y323 | DEPVADPYDQSFESR | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ELAVL1 | Q15717 | T118 | SGLPRTMTQKDVEDM | + |
| 4E-BP1 | Q13541 | T36 | PPGDYSTTPGGTLFS | |
| 4E-BP1 | Q13541 | T45 | GGTLFSTTPGGTRII | - |
| Akt1 (PKBa) | P31749 | S473 | RPHFPQFSYSASGTA | + |
| Akt1 (PKBa) | P31749 | T308 | KDGATMKTFCGTPEY | + |
| AR | P10275 | S650 | GEASSTTSPTEETTQ | - |
| ATF2 | P15336 | T69 | SVIVADQTPTPTRFL | + |
| ATF2 | P15336 | T71 | IVADQTPTPTRFLKN | + |
| Bcl-2 | P10415 | S70 | RDPVARTSPLQTPAA | |
| Bcl-2 | P10415 | S87 | AAAGPALSPVPPVVH | + |
| Bcl-2 | P10415 | T56 | FSSQPGHTPHPAASR | + |
| Bim | O43521 | S69 | GPLAPPASPGPFATR | |
| BTG2 | P78543 | S149 | LLGRSSPSKNYVMAV | |
| C-EBPa | P49715 | S21 | PMSSHLQSPPHAPSS | |
| Caldesmon | Q05682 | S789 | QSVDKVTSPTKV___ | |
| Caspase 3 | P42574 | S150 | FRGDRCRSLTGKPKL | - |
| Caspase 8 | Q14790 | S347 | FTGLKCPSLAGKPKV | - |
| CDX2 | Q99626 | S283 | RSVPEPLSPVSSLQA | |
| CHOP | P35638 | S79 | EVTSTSQSPHSPDSS | - |
| CHOP | P35638 | S82 | STSQSPHSPDSSQSS | - |
| Cip1 (p21, CDKN1A) | P38936 | S129 | SGEQAEGSPGGPGDS | - |
| Cip1 (p21, CDKN1A) | P38936 | T56 | NFDFVTETPLEGDFA | |
| cPLA2 | P47712 | S505 | LNTSYPLSPLSDFAT | + |
| CRYAB | P02511 | S59 | PSFLRAPSWFDTGLS | ? |
| Cx43 | P17302 | S278 | SSPTAPLSPMSPPGY | |
| Cx43 | P17302 | S281 | TAPLSPMSPPGYKLV | |
| Cyclin D1 (CCND1) | P24385 | T286 | EEVDLACTPTDVRDV | |
| Cyclin D2 (CCND2) | P30279 | T280 | DELDQASTPTDVRDI | |
| Cyclin D3 (CCND3) | P30281 | T283 | QGPSQTSTPTDVTAI | |
| DLX5 | P56178 | S217 | SDPMACNSPQSPAVW | |
| DLX5 | P56178 | S34 | MHHPSQESPTLPESS | |
| E2A | P15923 | S139 | LNSPGPLSPSGMKGT | |
| EEA1 | Q15075 | T1392 | CSAKNALTPSSKKPV | |
| EGFR | P00533 | T693 | RELVEPLTPSGEAPN | - |
| eIF2A | P05198 | S52 | MILLSELSRRRIRSI | - |
| Elk-1 | P19419 | S383 | IHFWSTLSPIAPRSP | + |
| ELK4 | P28324 | S381 | IHFWSTLSPVAPLSP | + |
| ELK4 | P28324 | S387 | LSPVAPLSPARLQGA | + |
| ERa (ESR1) | P03372 | S118 | LHPPPQLSPFLQPHG | + |
| ERa (ESR1) | P03372 | T311 | NSLALSLTADQMVSA | + |
| ETV6 | P41212 | S22 | ISYTPPESPVPSYAS | - |
| ETV6 | P41212 | S257 | ESHPKPSSPRQESTR | - |
| EWS | Q01844 | T79 | QPPTGYTTPTAPQAY | |
| FGFR1 | P11362 | S777 | SMPLDQYSPSFPDTR | ? |
| FOXO1A | Q12778 | S287 | GQEGAGDSPGSQFSK | - |
| FOXO1A | Q12778 | S295 | QFSKWPASPGSHSND | |
| FOXO1A | Q12778 | S329 | STISGRLSPIMTEQD | - |
| FOXO1A | Q12778 | S416 | PPNTSLNSPSPNYQK | + |
| FOXO1A | Q12778 | S432 | TYGQSSMSPLPQMPI | + |
| FOXO1A | Q12778 | S470 | KELLTSDSPPHNDIM | + |
| FOXO1A | Q12778 | T478 | PPHNDIMTPVDPGVA | |
| FOXO1A | Q12778 | T560 | NRLTQVKTPVQVPLP | |
| GR | P04150 | S211 | PGKETNESPWRSDLL | ? |
| H3.1 | P68431 | S10 | TKQTARKSTGGKAPR | |
| H3.2 | P84228 | S29 | ATKAARKSAPATGGV | |
| HBP1 | O14790 | S402 | GFSKNCGSPESSQLS | |
| HePTP | P35236 | S93 | ALQRQPPSPKQLEEE | - |
| HePTP | P35236 | T66 | EPICSVNTPREVTLH | - |
| HNF4A | P41235 | S167 | VLSRQITSPVSGING | |
| JDP-2 | Q8WYK2 | T148 | VRTDSVKTPESEGNP | |
| KHSRP | Q92945 | T692 | QAAYYGQTPVPGPQP | |
| KRT8 | P05787 | S432 | SAYGGLTSPGLSYSL | |
| KRT8 | P05787 | S74 | TVNQSLLSPLVLEVD | |
| Kv2.1 | Q14721 | S805 | ESSPLPTSPKFLRQN | |
| LIMK1 | P53667 | S310 | PGAGSLGSPASQRKD | |
| MAPKAPK2 | P49137 | S272 | SNHGLAISPGMKTRI | + |
| MAPKAPK2 | P49137 | T222 | TSHNSLTTPCYTPYY | + |
| MAPKAPK2 | P49137 | T25 | APPPQPPTPALPHPP | |
| MAPKAPK2 | P49137 | T334 | QSTKVPQTPLHTSRV | + |
| MAPKAPK3 | Q16644 | S251 | SNTGQAISPGMKRRI | |
| MAPKAPK3 | Q16644 | T201 | TTQNALQTPCYTPYY | + |
| MAPKAPK3 | Q16644 | T313 | QSMVVPQTPLHTARV | |
| MAPKAPK5 | Q8IW41 | T182 | IDQGDLMTPQFTPYY | + |
| MEF2A | Q02078 | S192 | TTLHRNVSPGAPQRP | |
| MEF2A | Q02078 | S223 | VPNGAGSSPVGNGFV | |
| MEF2A | Q02078 | S408 | SIKSEPISPPRDRMT | - |
| MEF2A | Q02078 | S479 | DPRGDFHSPIVLGRP | |
| MEF2A | Q02078 | S494 | PNTEDRESPSVKRMR | |
| MEF2A | Q02078 | S98 | KEHRGCDSPDPDTSY | |
| MEF2A | Q02078 | T108 | PDTSYVLTPHTEEKY | |
| MEF2A | Q02078 | T312 | QATQPLATPVVSVTT | + |
| MEF2A | Q02078 | T319 | TPVVSVTTPSLPPQG | + |
| MEF2C | Q06413 | T293 | QSAQSLATPVVSVAT | + |
| MEF2C | Q06413 | T300 | TPVVSVATPTLPGQG | + |
| MEF2C iso3 | Q06413 | S387 | TRHEAGRSPVDSLSS | + |
| MITF | O75030 | S414 | IPSTGLCSPDLVNRI | |
| MNK1 (MKNK1) | Q9BUB5 | T250 | NSCTPITTPELTTPC | + |
| MNK1 (MKNK1) | Q9BUB5 | T255 | ITTPELTTPCGSAEY | + |
| MNK2 (GPRK7) | Q9HBH9 | T244 | GDCSPISTPELLTPC | + |
| MNK2 (GPRK7) | Q9HBH9 | T249 | ISTPELLTPCGSAEY | + |
| MNK2 (GPRK7) | Q9HBH9 | T379 | APENTLPTPMVLQRN | + |
| MRP14 | P06702 | T113 | KPGLGEGTP______ | |
| MSK1 (RPS6KA5) | O75582 | S360 | TEMDPTYSPAALPQS | + |
| MSK1 (RPS6KA5) | O75582 | T581 | PDNQPLKTPCFTLHY | + |
| MSK1 (RPS6KA5) | O75582 | T700 | LSSNPLMTPDILGSS | + |
| MSK2 (RPS6KA4) | O75676 | S196 | EEKERTFSFCGTIEY | + |
| MSK2 (RPS6KA4) | O75676 | S343 | TRLEPVYSPPGSPPP | + |
| MSK2 (RPS6KA4) | O75676 | S347 | PVYSPPGSPPPGDPR | + |
| MSK2 (RPS6KA4) | O75676 | S360 | PRIFQGYSFVAPSIL | + |
| MSK2 (RPS6KA4) | O75676 | T568 | SPGVPMQTPCFTLQY | + |
| MSK2 (RPS6KA4) | O75676 | T687 | RSSPPLRTPDVLESS | + |
| Myf-6 | P23409 | S31 | PLEVAEGSPLYPGSD | |
| Myf-6 | P23409 | S42 | PGSDGTLSPCQDQMP | |
| MZF-1 | P28698 | S256 | EEAGGIFSPGFALQL | |
| MZF-1 | P28698 | S294 | GLSGQIQSPSREGGF | |
| MZF-1 iso2 | P28698 | S257 | EEVGGIFSPGFTLQM | |
| MZF-1 iso2 | P28698 | S275 | TAEPDMMSPHLHVPW | |
| MZF-1 iso2 | P28698 | S295 | SLSGQIQSPTTEGGF | |
| NCoA2 | Q15596 | S736 | TIKQEPVSPKKKENA | |
| NCOA3 (SRC-3) | Q9Y6Q9 | S505 | SPVAGVHSPMASSGN | - |
| NCOA3 (SRC-3) | Q9Y6Q9 | S543 | SLLSTLSSPGPKLDN | - |
| NCOA3 (SRC-3) | Q9Y6Q9 | S860 | NRAVSLDSPVSVGSS | - |
| NCOA3 (SRC-3) | Q9Y6Q9 | S867 | SPVSVGSSPPVKNIS | - |
| NFAT2 | O95644 | S172 | YRDPSCLSPASSLSS | |
| NFAT3 | Q14934 | S168 | QGGGAFFSPSPGSSS | - |
| NFAT3 | Q14934 | S170 | GGAFFSPSPGSSSLS | - |
| p38a MAPK (MAPK14) | Q16539 | T180 | RHTDDEMTGYVATRW | + |
| p38a MAPK (MAPK14) | Q16539 | Y182 | TDDEMTGYVATRWYR | + |
| p47phox | P14598 | S345 | QARPGPQSPGSPLEE | + |
| p47phox | P14598 | S348 | PGPQSPGSPLEEERQ | |
| p53 | P04637 | S15 | PSVEPPLSQETFSDL | + |
| p53 | P04637 | S33 | LPENNVLSPLPSQAM | + |
| p53 | P04637 | S46 | AMDDLMLSPDDIEQW | + |
| PAK6 | Q9NQU5 | S165 | MPWPEPQSPRVLPNG | + |
| PDX1 | P52945 | S61 | LGALEQGSPPDISPY | + |
| PDX1 | P52945 | S66 | QGSPPDISPYEVPPL | + |
| PGC-1 | Q9UBK2 | S266 | SLPLTPESPNDPKGS | |
| PGC-1 | Q9UBK2 | T263 | TTLSLPLTPESPNDP | |
| PGC-1 | Q9UBK2 | T299 | AGLTPPTTPPHKANQ | |
| PIP5K2B | P78356 | S326 | SYGTPPDSPGNLLSF | - |
| PKCe (PRKCE) | Q02156 | S350 | RSKSAPTSPCDQEIK | |
| PKR (PRKR; EIF2AK2) | P19525 | T451 | KRTRSKGTLRYMSPE | - |
| PPARA | Q07869 | S12 | ESPLCPLSPLEAGDL | - |
| PPARA | Q07869 | S211 | LEAGDLESPLSEEFL | |
| PPARA | Q07869 | S6 | __MVDTESPLCPLSP | - |
| PXN | P49023 | S83 | FIHQQPQSSSPVYGS | |
| PXN iso2 | P49023 | S85 | HQQPQSSSPVYGSSA | |
| RARG2 | P13631 | S77 | SEEMVPSSPSPPPPP | + |
| RARG2 | P13631 | S79 | EMVPSSPSPPPPPRV | + |
| SCN10A | Q9Y5Y9 | S552 | QQGPLPRSPLPQPSN | |
| SCN8A | Q9UQD0 | S553 | SLLSIPGSPFLSRHN | |
| Shc1 | P29353 | S36 | TPPEELPSPSASSLG | - |
| Shc1 | P29353 | S54 | PPLPGDDSPTTLCSF | + |
| Shc1 | P29353 | T386 | PTAPNAQTPSHLGAT | + |
| SIAH2 | O43255 | S28 | PQPQHTPSPAAPPAA | |
| SLC9A1 (NHE1) | P19634 | S723 | VITIDPASPQSPESV | |
| SLC9A1 (NHE1) | P19634 | S726 | IDPASPQSPESVDLV | + |
| SLC9A1 (NHE1) | P19634 | S729 | ASPQSPESVDLVNEE | + |
| SLC9A1 (NHE1) | P19634 | T718 | KEDLPVITIDPASPQ | |
| STAT1 | P42224 | S727 | TDNLLPMSPEEFDEV | - |
| STAT3 | P40763 | S727 | NTIDLPMSPRTLDSL | - |
| STAT4 | Q14765 | S721 | PSDLLPMSPSVYAVL | |
| STMN1 | P16949 | S25 | QAFELILSPRSKESV | |
| STMN1 | P16949 | S38 | SVPEFPLSPPKKKDL | |
| STMN2 | Q93045 | S62 | ELILKPPSPISEAPR | |
| STMN2 | Q93045 | S73 | EAPRILASPKKKDLS | |
| TAB1 | Q15750 | S423 | QMVNGAHSASTLDEA | |
| TAB1 | Q15750 | S438 | TPTLTNQSPTLTLQS | |
| TAB1 | Q15750 | T431 | ASTLDEATPTLTNQS | |
| TAB2 | Q9NYJ8 | S423 | SAASRNMSGQVSMGP | |
| TAB2 | Q9NYJ8 | T431 | ASTLDEATPTLTNQS | |
| Tau iso5 (Tau-C) | P10636-5 | S185 | APKTPPSSGEPPKSG | |
| Tau iso5 (Tau-C) | P10636-5 | S202 | SGYSSPGSPGTPGSR | |
| Tau iso5 (Tau-C) | P10636-5 | T205 | SSPGSPGTPGSRSRT | |
| Tau iso5 (Tau-C) | P10636-5 | T245 | SAKSRLQTAPVPMPD | |
| Tau iso9 (Tau-F) | P10636-9 | S305 | KHVPGGGSVQIVYKP | |
| Tau iso9 (Tau-F) | P10636-9 | S356 | RVQSKIGSLDNITHV | |
| Tau iso9 (Tau-F) | P10636-9 | S404 | PVVSGDTSPRHLSNV | |
| USF1 | P22415 | T153 | EALLGQATPPGTGQF | |
| ZFYVE20 | Q9H1K0 | S215 | ESLSTHTSPSQSPNS |
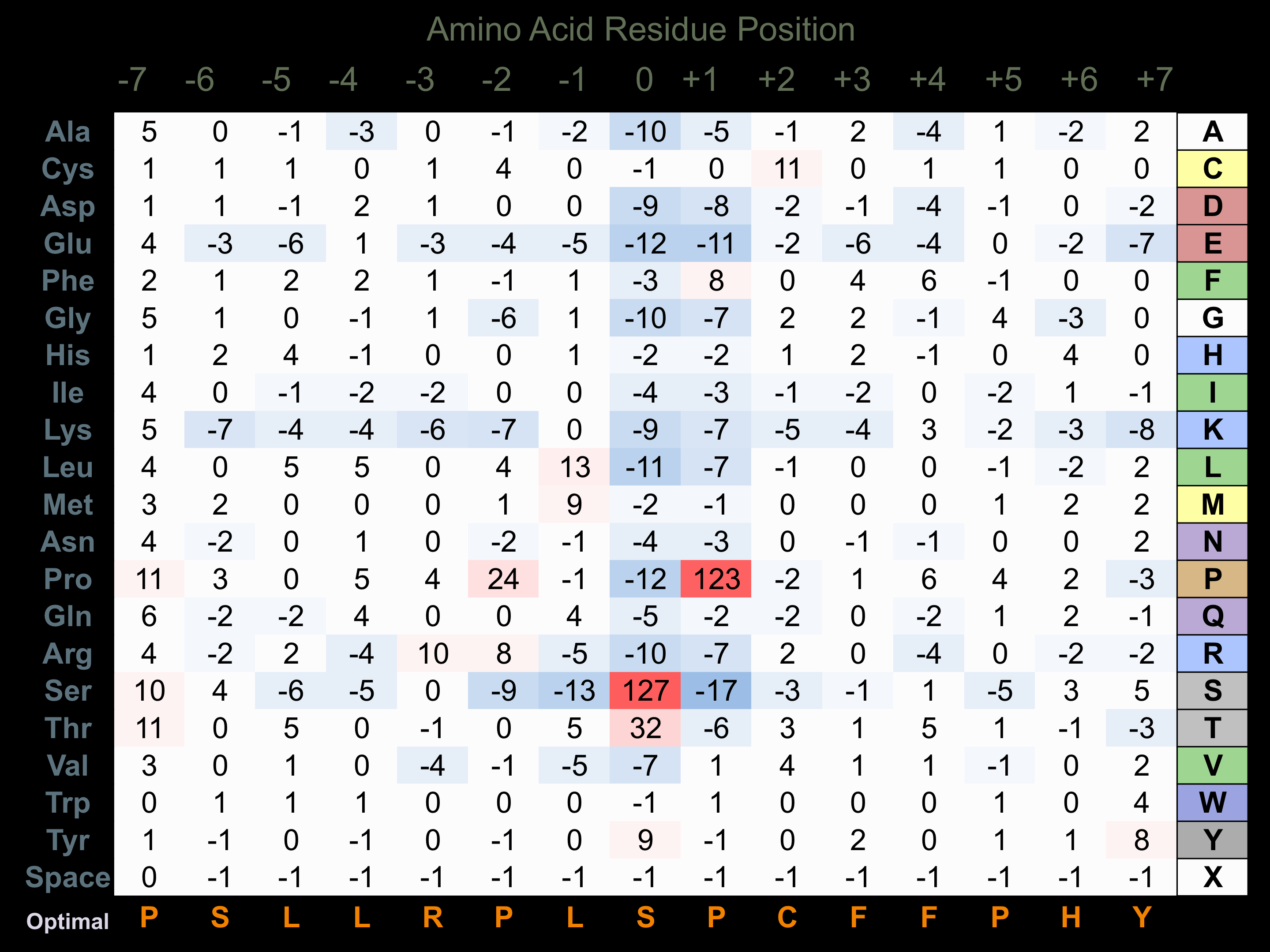
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
Matrix Type:
Experimentally derived from alignment of 173 known protein substrate phosphosites and 55 peptides phosphorylated by recombinant p38a MAPK in vitro tested in-house by Kinexus. Note that additional binding sites on p38 substrates with D motifs (consensus= K/R-k-x-s-l/p-l-l-l-p-p or p-x-L/v/i-x-p-p-x-x-x-x-l-l-x-r/k-k/r-R/k-K/r) facilitate higher selectivity for phosphorylation by this protein kinase.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Infectious disease, and neuronal disorders
Specific Diseases (Non-cancerous):
Influenza; Alexander disease
Comments:
p38a is implicated in the pathophysiology of inflammatory bowel disease. It may be a potential therapeutic target for spinal muscle atrophy, which is an autosomal recessive neurodegenerative disease characterized by progressive muscle atrophy. Spinal muscle atrophy results from deletion or mutation of SMN1 gene, which can be induced by p38a activitation.
Comments:
p38a may be also involved in drug resistance development of acute myeloid leukemias.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= +221, p<0.012); and Brain oligodendrogliomas (%CFC= +193, p<0.06). The COSMIC website notes an up-regulated expression score for p38a in diverse human cancers of 565, which is 1.2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 152 for this protein kinase in human cancers was 2.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25411 diverse cancer specimens. This rate is very similar (+ 6% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.48 % in 805 skin cancers tested; 0.38 % in 1093 large intestine cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,694 cancer specimens
Comments:
Only 2 deletions, 3 insertions, and no complex mutations are noted on the COSMIC website.

