Nomenclature
Short Name:
ABL
Full Name:
Abelson murine leukemia viral oncogene homologue 1
Alias:
- Abelson murine leukemia viral oncogene 1
- P150
- V-abl Abelson murine leukemia viral oncogene 1
- ABL1
- C-ABL
- EC 2.7.10.2
- JTK7
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Abl
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
122,873
# Amino Acids:
1130
# mRNA Isoforms:
2
mRNA Isoforms:
124,955 Da (1149 AA; P00519-2); 122,873 Da (1130 AA; P00519)
4D Structure:
Interacts with SORBS1 following insulin stimulation. Found in a trimolecular complex containing CDK5 and CABLES1. Interacts with CABLES1 and PSTPIP1. Interacts with ZDHHC16 By similarity. Interacts with INPPL1/SHIP2. Interacts with the 14-3-3 proteins, YWHAB, YWHAE, YWHAG, YWHAH, SFN AND YWHAZ; the interaction with 14-3-3 proteins requires phosphorylation on Thr-735 and, sequesters ABL1 into the cytoplasm. Interacts with PSMA7
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
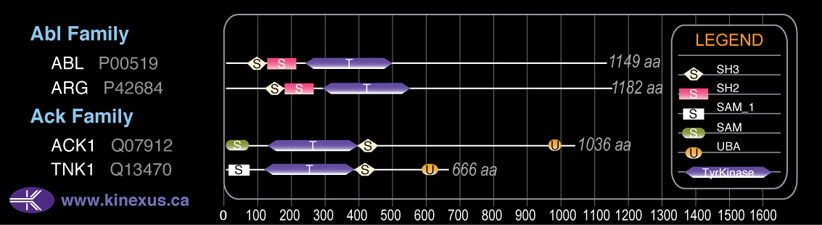
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K274, K711 (N6), K759, .
Myristoylated:
G2 (in P00519-2 isoform).
Serine phosphorylated:
S43, S50, S75, S113, S142, S180, S187, S229, S265, S417, S446+, S535, S559, S569, S601, S618+, S619+, S620, S642, S659, S676, S679, S683, S690, S696, S708, S709, S710, S718, S805, S808, S809, S855, S859, S884, S917, S919, S936, S943, S949, S977, S1106.
Threonine phosphorylated:
T117, T178, T224, T243, T267, T306, T389, T392, T394+, T532, T597, T610, T613, T649, T653, T692, T700, T735+, T751, T781, T814, T823, T844, T852, T854, T935, T963, T1111Y70+.
Tyrosine phosphorylated:
Y70+, Y93, Y115, Y128, Y139+, Y167, Y172, Y174, Y185, Y215, Y226+, Y232, Y253-, Y257-, Y264-, Y312+, Y320, Y393+, Y413, Y456, Y469, Y1064, Y1070.
Ubiquitinated:
K285.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 15
15
921
19
1028
 4
4
278
18
120
 5
5
335
21
169
 9
9
549
99
740
 17
17
1062
14
813
 12
12
730
56
1588
 15
15
926
39
607
 10
10
617
69
1389
 9
9
579
10
354
 4
4
248
135
214
 2
2
116
54
99
 7
7
453
231
592
 2
2
145
32
72
 7
7
412
19
229
 3
3
183
50
170
 5
5
288
12
121
 4
4
270
611
797
 3
3
204
35
124
 2
2
134
100
85
 13
13
791
56
709
 6
6
355
50
413
 4
4
258
53
324
 3
3
199
30
93
 2
2
136
34
92
 4
4
251
50
324
 10
10
630
66
758
 2
2
146
35
61
 3
3
192
34
136
 6
6
384
34
296
 2
2
133
14
77
 13
13
819
30
492
 100
100
6230
21
9941
 19
19
1172
54
1139
 15
15
935
31
848
 3
3
157
40
178
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 97.4
97.4
97.7
100 78.8
78.8
81.2
- -
-
-
91 -
-
-
97 91.5
91.5
93.8
92 -
-
-
- 87.7
87.7
91.2
89 87.6
87.6
91
89 -
-
-
- 81.7
81.7
86.3
- 83.6
83.6
88.4
84 54.3
54.3
65.5
75 69.9
69.9
77.3
76 -
-
-
- 36.2
36.2
47
- -
-
-
- 33.2
33.2
48.9
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CRK - P46108 |
| 2 | BCR - P11274 |
| 3 | ABI2 - Q9NYB9 |
| 4 | RB1 - P06400 |
| 5 | CBL - P22681 |
| 6 | DOK1 - Q99704 |
| 7 | NCK1 - P16333 |
| 8 | PIK3R1 - P27986 |
| 9 | RIN1 - Q13671 |
| 10 | HCK - P08631 |
| 11 | TP53 - P04637 |
| 12 | BCAR1 - P56945 |
| 13 | PTPN6 - P29350 |
| 14 | MAP4K1 - Q92918 |
| 15 | BIN1 - O00499 |
Regulation
Activation:
Stabilized in the inactive form by an association between the SH3 domain and the SH2-TK linker region, interactions of the amino-terminal cap, and contributions from an amino-terminal myristoyl group and phospholipids. Activated by autophosphorylation as well as by SRC-family kinase-mediated phosphorylation. Activated by RIN1 binding to the SH2 and SH3 domains.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
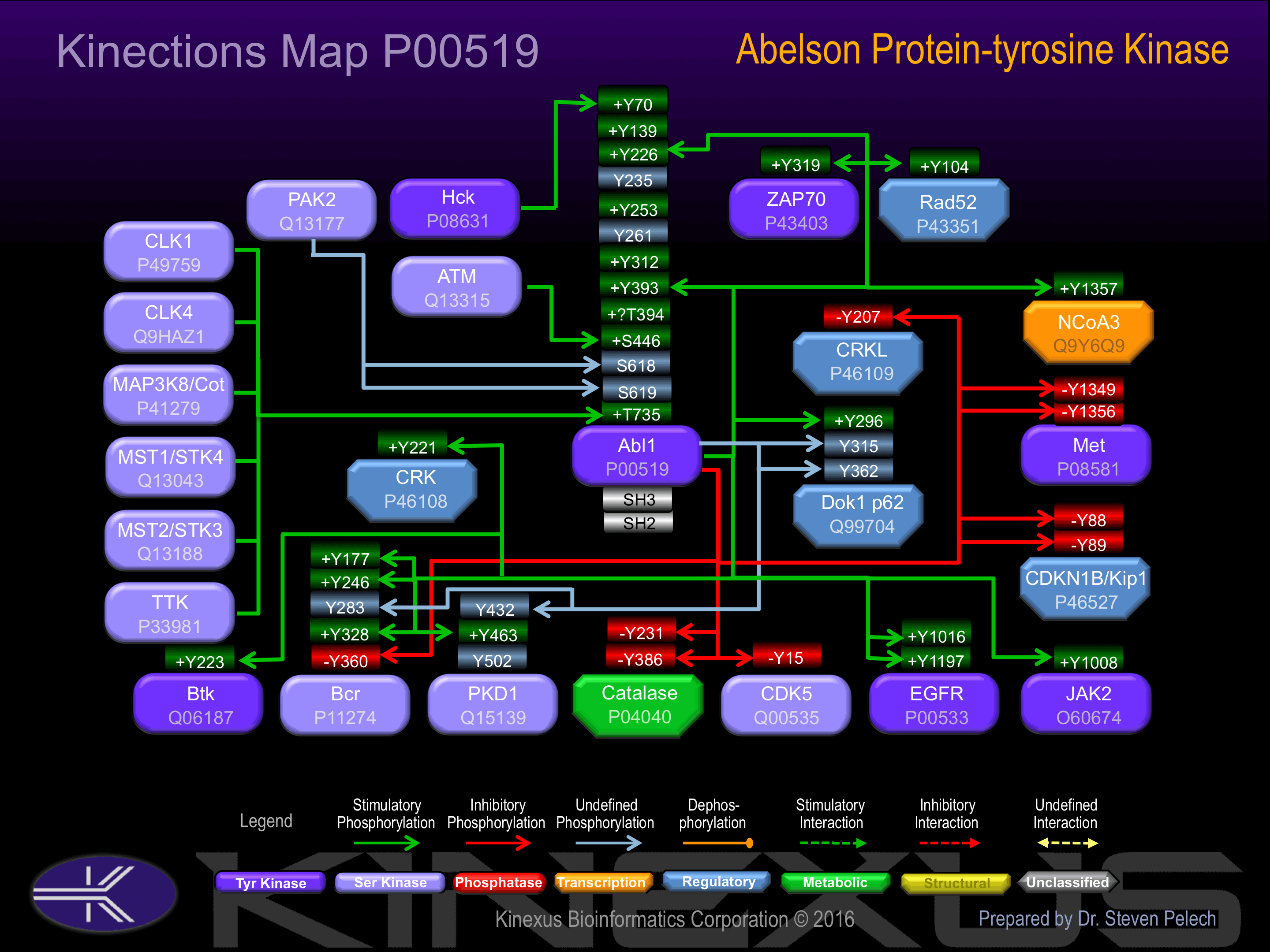
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| LYN | P07948 | Y70 | PNLFVALYDFVASGD | + |
| FYN | P06241 | Y70 | PNLFVALYDFVASGD | + |
| HCK | P08631 | Y70 | PNLFVALYDFVASGD | + |
| LYN | P07948 | Y115 | QGWVPSNYITPVNSL | |
| FYN | P06241 | Y115 | QGWVPSNYITPVNSL | |
| HCK | P08631 | Y115 | QGWVPSNYITPVNSL | |
| LYN | P07948 | Y128 | SLEKHSWYHGPVSRN | |
| FYN | P06241 | Y128 | SLEKHSWYHGPVSRN | |
| HCK | P08631 | Y128 | SLEKHSWYHGPVSRN | |
| LYN | P07948 | Y139 | VSRNAAEYLLSSGIN | + |
| FYN | P06241 | Y139 | VSRNAAEYLLSSGIN | + |
| HCK | P08631 | Y139 | VSRNAAEYLLSSGIN | + |
| LYN | P07948 | Y172 | LRYEGRVYHYRINTA | |
| FYN | P06241 | Y172 | LRYEGRVYHYRINTA | |
| HCK | P08631 | Y172 | LRYEGRVYHYRINTA | |
| LYN | P07948 | Y185 | TASDGKLYVSSESRF | |
| FYN | P06241 | Y185 | TASDGKLYVSSESRF | |
| HCK | P08631 | Y185 | TASDGKLYVSSESRF | |
| LYN | P07948 | Y215 | GLITTLHYPAPKRNK | |
| FYN | P06241 | Y215 | GLITTLHYPAPKRNK | |
| HCK | P08631 | Y215 | GLITTLHYPAPKRNK | |
| LYN | P07948 | Y226 | KRNKPTVYGVSPNYD | + |
| FYN | P06241 | Y226 | KRNKPTVYGVSPNYD | + |
| ABL | P00519 | Y226 | KRNKPTVYGVSPNYD | + |
| HCK | P08631 | Y226 | KRNKPTVYGVSPNYD | + |
| LYN | P07948 | Y393 | RLMTGDTYTAHAGAK | + |
| FYN | P06241 | Y393 | RLMTGDTYTAHAGAK | + |
| ABL | P00519 | Y393 | RLMTGDTYTAHAGAK | + |
| HCK | P08631 | Y393 | RLMTGDTYTAHAGAK | + |
| ATM | Q13315 | S446 | PYPGIDLSQVYELLE | + |
| CDK1 | P06493 | S569 | LDHEPAVSPLLPRKE | |
| PAK2 | Q13177 | S618 | APTPPKRSSSFREMD | + |
| PAK2 | Q13177 | S619 | PTPPKRSSSFREMDG | + |
| MST2 | Q13188 | T735 | DTEWRSVTLPRDLQS | + |
| MST1 | Q13043 | T735 | DTEWRSVTLPRDLQS | + |
| TTK | P33981 | T735 | DTEWRSVTLPRDLQS | + |
| CLK1 | P49759 | T735 | DTEWRSVTLPRDLQS | + |
| CLK4 | Q9HAZ1 | T735 | DTEWRSVTLPRDLQS | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Abi-1 | Q8IZP0 | Y213 | PPTVPNDYMTSPARL | |
| Abl1 | P00519 | Y226 | KRNKPTVYGVSPNYD | + |
| Abl1 | P00519 | Y393 | RLMTGDTYTAHAGAK | + |
| Abl1 iso2 | P00519-2 | Y245 | KRNKPTVYGVSPNYD | + |
| Abl1 iso2 | P00519-2 | Y412 | RLMTGDTYTAHAGAK | + |
| ANXA1 | P04083 | Y20 | IENEEQEYVQTVKSS | |
| APP | P05067 | Y757 | SKMQQNGYENPTYKF | |
| Bcr | P11274 | Y177 | ADAEKPFYVNVEFHH | + |
| Bcr | P11274 | Y246 | SCGVDGDYEDAELNP | + |
| Bcr | P11274 | Y283 | YQPYQSIYVGGMMEG | |
| Bcr | P11274 | Y328 | FEDCGGGYTPDCSSN | + |
| Bcr | P11274 | Y360 | VSPSPTTYRMFRDKS | - |
| Btk | Q06187 | Y223 | LKKVVALYDYMPMNA | + |
| Caspase 9 | P55211 | Y153 | RGNADLAYILSMEPC | |
| Catalase | P04040 | Y230 | NANGEAVYCKFHYKT | - |
| Catalase | P04040 | Y385 | YRARVANYQRDGPMC | - |
| Cbl | P22681 | Y700 | EGEEDTEYMTPSSRP | |
| Cbl | P22681 | Y774 | SENEDDGYDVPKPPV | |
| CD19 | P15391 | Y508 | EDMRGILYAAPQLRS | |
| CDK5 | Q00535 | Y15 | EKIGEGTYGTVFKAK | - |
| CDV-3 | Q9UKY7 | Y95 | LEQKEVDYSGLRVQA | |
| Crk | P46108 | Y221 | GGPEPGPYAQPSVNT | + |
| CrkL | P46109 | Y207 | IPEPAHAYAQPQTTT | - |
| CTNNB1 | P35222 | Y489 | QNAVRLHYGLPVVVK | |
| CTNNB1 | P35222 | Y654 | RNEGVATYAAAVLFR | + |
| CTNNB1 | P35222 | Y86 | VADIDGQYAMTRAQR | |
| DDX5 | P17844 | Y593 | NGMNQQAYAYPATAA | |
| Dok1 p62 | Q99704 | Y296 | LDSPPALYAEPLDSL | + |
| Dok1 p62 | Q99704 | Y315 | CPSQDSLYSDPLDST | ? |
| Dok1 p62 | Q99704 | Y362 | DPKEDPIYDEPEGLA | + |
| EGFR | P00533 | Y1016 | DVVDADEYLIPQQGF | + |
| EGFR | P00533 | Y1197 | STAENAEYLRVAPQS | + |
| ERCC6 | Q03468 | Y932 | GANRVVIYDPDWNPS | |
| Fe65 | O00213 | Y547 | VQKFQVYYLGNVPVA | |
| GPX1 | P07203 | Y96 | EILNSLKYVRPGGGF | |
| IkBa | P25963 | Y305 | FTEDELPYDDCVFGG | - |
| JAK2 | O60674 | Y1008 | LPQDKEYYKVKEPGE | + |
| Jun (c-Jun) | P05412 | Y170 | LHSEPPVYANLSNFN | |
| Lasp-1 | Q14847 | Y171 | IPTSAPVYQQPQQQP | |
| MDM2 | Q00987 | Y276 | SDEDDEVYQVTVYQA | |
| MDM2 | Q00987 | Y394 | QSQESEDYSQPSTSS | |
| Mena | Q8N8S7 | S22 | PTLPRQNSQLPAQVQ | |
| Met | P08581 | Y1349 | STFIGEHYVHVNATY | - |
| Met | P08581 | Y1356 | YVHVNATYVNVKCVA | - |
| MYLK1 (smMLCK) | Q15746 | Y1449 | EKEPEVDYRTVTINT | |
| MYLK1 (smMLCK) | Q15746 | Y1575 | QISEGVEYIHKQGIV | |
| MYLK1 (smMLCK) | Q15746 | Y1635 | VAPEVINYEPIGYAT | |
| MYLK1 (smMLCK) | Q15746 | Y231 | NQDDVGVYTCLVVNG | |
| MYLK1 (smMLCK) | Q15746 | Y464 | QEGSIEVYEDAGSHY | + |
| MYLK1 (smMLCK) | Q15746 | Y556 | LNGQPIQYARSTCEA | |
| MYLK1 (smMLCK) | Q15746 | Y611 | KSSRkSEYLLPVAPS | |
| MYLK1 (smMLCK) | Q15746 | Y792 | QPWHAGQYEILLKNR | |
| MYLK1 (smMLCK) | Q15746 | Y846 | DGGGSDRYGSLRPGW | |
| MyoD | P15172 | Y212 | CSDGMMDYSGPPSGA | |
| MyoD | P15172 | Y30 | FATTDDFYDDPCFDS | |
| NCoA3 | Q9Y6Q9 | Y1357 | HPQAASIYQSSEMKG | ? |
| p27Kip1 | P46527 | Y88 | KGSLPEFYYRPPRPP | - |
| p27Kip1 | P46527 | Y89 | GSLPEFYYRPPRPPK | - |
| p73 | O15350 | Y99 | SVPTHSPYAQPSSTF | |
| PIK3R1 | P27986 | Y368 | STKMHGDYTLTLRKG | |
| PKD1 (PRKCM) | Q15139 | Y432 | KEGWMVHYTSKDTLR | |
| PKD1 (PRKCM) | Q15139 | Y463 | NDTGSRYYKEIPLSE | + |
| PKD1 (PRKCM) | Q15139 | Y502 | TTANVVYYVGENVVN | |
| PLSCR1 | O15162 | Y69 | PVPNQPVYNQPVYNQ | |
| PLSCR1 | O15162 | Y74 | PVYNQPVYNQPVGAA | |
| PPM1B | O75688 | S186 | RIQNAGGSVMIQRVN | |
| PPM1B | O75688 | Y367 | RNVIEAVYSRLNPHR | |
| PSMA7 | O14818 | Y153 | QTDPSGTYHAWKANA | |
| PXN | P49023 | Y118 | VGEEEHVYSFPNKQK | |
| PXN | P49023 | Y31 | FLSEETPYSYPTGNH | |
| Rad51 | Q06609 | Y315 | ETRICKIYDSPCLPE | |
| Rad51 | Q06609 | Y54 | HTVEAVAYAPKKELI | |
| Rad52 | P43351 | Y104 | DLNNGKFYVGVCAFV | + |
| Rad9 | Q99638 | Y28 | SRIGDELYLEPLEDG | ? |
| Rb | P06400 | Y805 | RIPGGNIYISPLKSP | |
| TOP1 | P11387 | Y268 | AKMLDHEYTTKEIFR | |
| Vinexin | O60504 | Y127 | QHPDPAWYQTWPGPG | |
| WASF2 | Q9Y6W5 | Y150 | GKEALKFYTDFSYFF | |
| WASP | P42768 | Y291 | AETSKLIYDFIEDQG | + |
| YAP1 | P46937 | Y357 | SGLSMSSYSVPRTPD | |
| ZAP70 | P43403 | Y319 | TSVYESPYSDPEELK | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
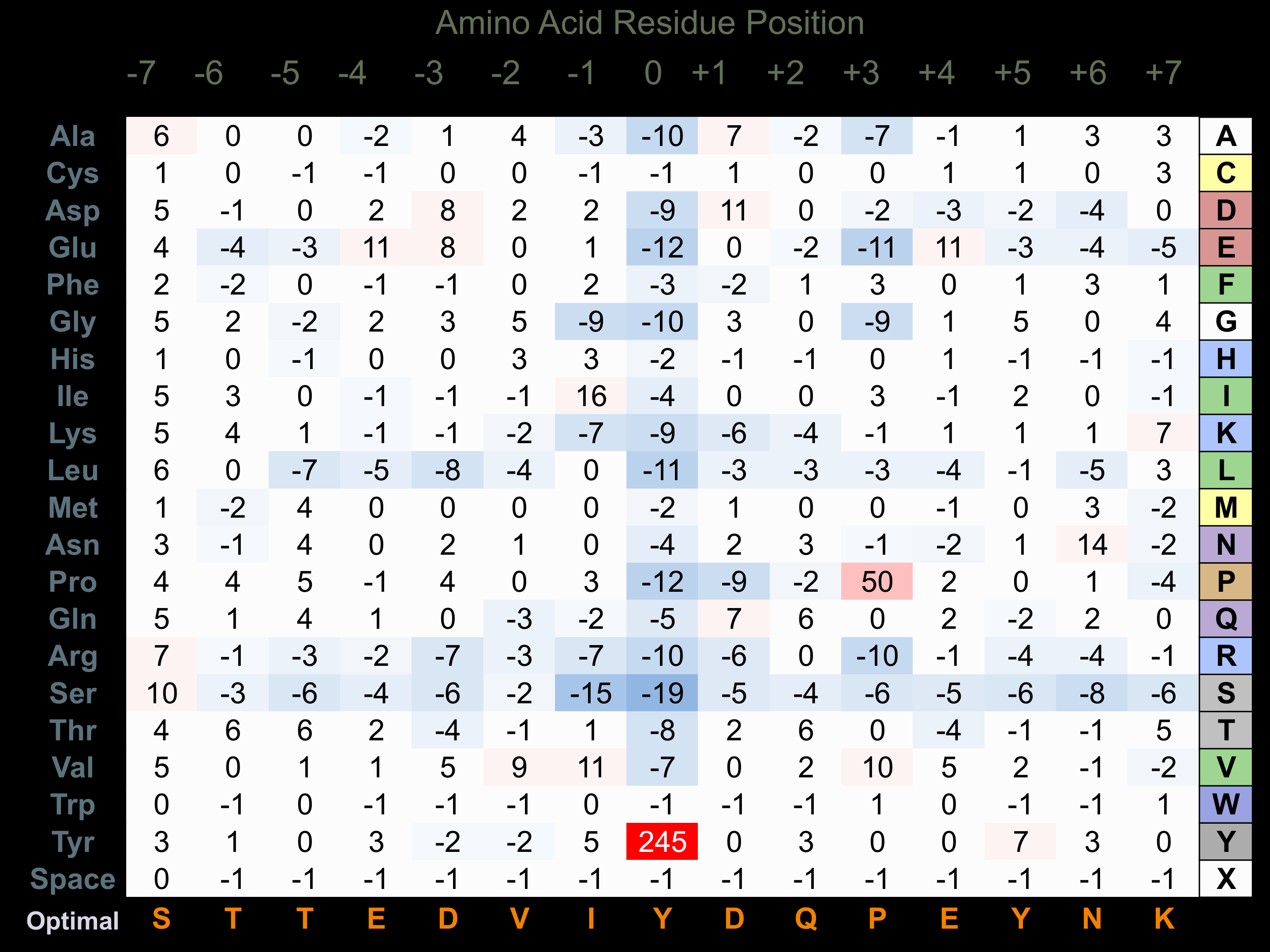
Matrix Type:
Experimentally derived from alignment of 155 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Chronic myeloid leukemia (CML); Precursor B-cell acute lymphoblastic leukemia; Precursor T-cell acute lymphoblastic leukemia;
Comments:
ABL1 is a known oncoprotein (OP), although the wild-type form of ABL1 may be a tumour-suppressor protein. Cancer-related mutations in human tumours point to a gain of function of the protein kinase. The active form of the protein kinase normally acts to promote tumour cell proliferation. Translocation t(9;22)(q34;q11)result in chimeric proteins from BCR and ABL1, and this is a cause of chronic myeloid leukemia (CML). The translocation also produces a BCR-ABL found in acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL). BCR-ABL fusion protein results in constitutively active phosphotransferase activity by inhibiting 3BP1/ABL interaction. BCR-ABL induces the Ras, PI3K, and Myc cell proliferation pathways. The most common mutations are clustered within the kinase catalytic domain. T315 is near kinase Subdomain III. G250 and E255 are located around the ATP-binding kinase Subdomain I. It is likely that these common mutations may inactive Abl1's catalytic activity. BCR-Abl1 is found in the cytoplasm and nucleus. The phosphorylation of cytoplasmic proteins and receptors in the plasma membrane may result in a gain of function of Abl1 to stimulate cell proliferation.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= -64, p<0.014); Large B-cell lymphomas (%CFC= +84, p<0.009); Malignant pleural mesotheliomas (MPM) tumours (%CFC= +76, p<0.059); Ovary adenocarcinomas (%CFC= +55, p<0.059); Prostate cancer - primary (%CFC= +192, p<0.0001); Skin fibrosarcomas (%CFC= +83); Skin melanomas - malignant (%CFC= +171, p<0.0001); T-cell prolymphocytic leukemia (%CFC= +54, p<0.035); and Uterine leiomyomas (%CFC= -59, p<0.056). The COSMIC website notes an up-regulated expression score for ABL1 in diverse human cancers of 322, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 192 for this protein kinase in human cancers was 3.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.4 % in 34454 diverse cancer specimens. This rate is 5.3-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 1.49 % in 7546 haematopoietic and lymphoid cancers tested; 0.4 % in 1490 large intestine cancers tested; 0.26 % in 1100 skin cancers tested; 0.19 % in 708 endometrium cancers tested; 0.13 % in 856 stomach cancers tested; 0.09 % in 1106 ovary cancers tested; 0.08 % in 2357 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: T315I (302); G250E (133); E255K (116); E255V (29); Y253H (103); Y253F (16); M351T (100); M244V (90); F317L (82); F359V (71); H396R (41); E355G (36); Q252H (30); E459K (24).
Comments:
Most point mutations are located within the protein kinase catalytic domain.