Nomenclature
Short Name:
ASK1
Full Name:
Mitogen-activated protein kinase kinase kinase 5
Alias:
- Apoptosis signal regulating kinase 1
- MAP3K5
- MAPK/ERK kinase kinase 5
- MAPKKK5
- MEK kinase 5
- MEKK5
- Apoptosis signal-regulating kinase 1
- ASK-1
- Kinase ASK1
- M3K5
Classification
Type:
Protein-serine/threonine kinase
Group:
STE
Family:
STE11
SubFamily:
NA
Structure
Mol. Mass (Da):
154,537
# Amino Acids:
1374
# mRNA Isoforms:
2
mRNA Isoforms:
154,537 Da (1374 AA; Q99683); 69,839 Da (621 AA; Q99683-2)
4D Structure:
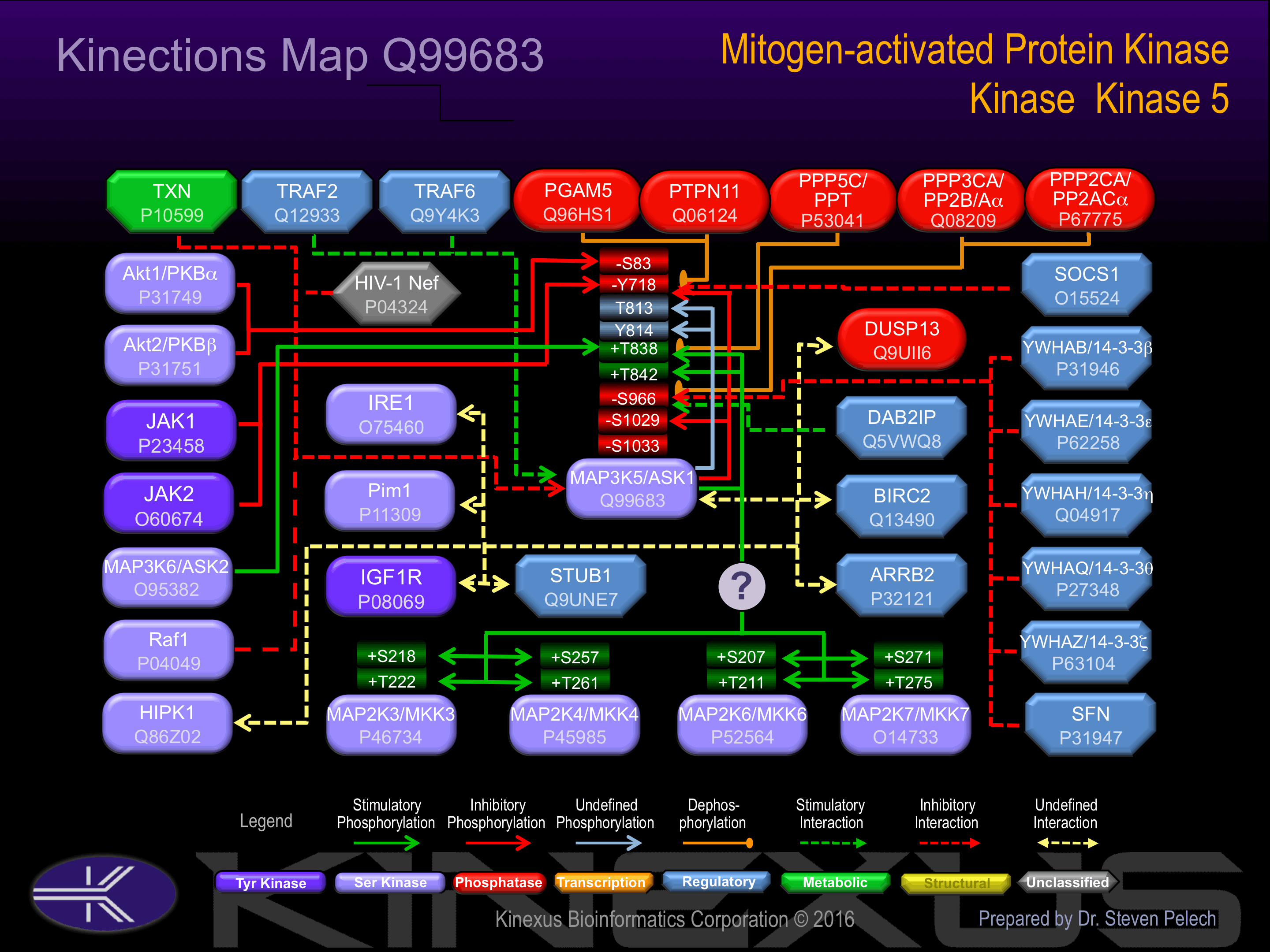
Homodimer when inactive. Binds both upstream activators and downstream substrates in multimolecular complexes. Associates with and inhibited by HIV-1 Nef. Interacts with DAB2IP and PPM1L. Interacts with ARRB2.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
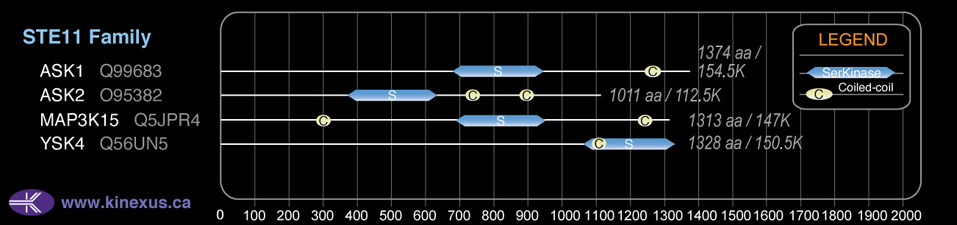
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 680 | 938 | Pkinase |
| 1244 | 1285 | Coiled-coil |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K946.
Serine phosphorylated:
S82, S83-, S115, S461, S821, S958, S964, S966-, S986, S1029-, S1033, S1240.
Sumoylated:
K535, K1083, K1114.
Threonine phosphorylated:
T528, T813+, T825, T838+, T842-, T1109, T1326.
Tyrosine phosphorylated:
Y570, Y574, Y718-, Y798, Y814, Y982.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 75
75
818
29
829
 49
49
529
15
155
 9
9
101
30
62
 39
39
428
113
453
 76
76
823
25
599
 14
14
152
82
167
 21
21
232
31
359
 44
44
483
63
841
 65
65
703
17
526
 11
11
116
100
83
 7
7
81
49
79
 58
58
635
191
691
 11
11
118
52
59
 29
29
317
12
178
 19
19
204
43
130
 83
83
900
15
553
 15
15
160
140
138
 6
6
69
38
62
 7
7
75
108
45
 67
67
727
109
741
 21
21
233
41
122
 13
13
147
45
96
 14
14
152
40
94
 10
10
110
39
66
 9
9
97
41
86
 46
46
500
79
661
 11
11
121
55
58
 9
9
98
39
55
 9
9
99
39
67
 5
5
59
28
44
 82
82
898
24
756
 55
55
598
36
591
 20
20
219
72
391
 100
100
1089
57
860
 48
48
523
35
1746
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.3
99.3
99.4
100 44.9
44.9
61.4
99 -
-
-
96 -
-
-
97 68
68
73.1
97 -
-
-
- 94.2
94.2
95.8
95 92.7
92.7
94.4
94 -
-
-
- 84.4
84.4
87.8
- 80.4
80.4
87
89 -
-
-
77 71.8
71.8
84.1
74 -
-
-
- 40.7
40.7
58.2
- -
-
-
- -
-
-
- 44.7
44.7
60.8
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | YWHAZ - P63104 |
| 2 | PDCD6 - O75340 |
| 3 | GADD45B - O75293 |
| 4 | IGF1R - P08069 |
| 5 | ERN1 - O75460 |
| 6 | AHRR - A9YTQ3 |
| 7 | AKT1 - P31749 |
| 8 | RAF1 - P04049 |
| 9 | QARS - P47897 |
| 10 | CDC25A - P30304 |
| 11 | RB1CC1 - Q8TDY2 |
| 12 | YWHAE - P62258 |
| 13 | YWHAH - Q04917 |
| 14 | GSTP1 - P09211 |
| 15 | MAPK8IP3 - Q9UPT6 |
Regulation
Activation:
Activated by phosphorylation at Thr-838 ; Binds to, and stabilizes MAP3K6 and is activated by MAP3K6 by phosphorylation on Thr-838. ; Dephosphorylated and activated by PGAM5.
Inhibition:
Contains an N-terminal autoinhibitory domain. ; inhibited by phosphorylation at Ser-966 and Ser-1033. ; Inhibited by HIV-1 Nef.
Synthesis:
By TNF-alpha.
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| AKT3 | Q9Y243 | S83 | ATRGRGSSVGGGSRR | - |
| Akt2 | P31751 | S83 | ATRGRGSSVGGGSRR | - |
| Akt1 | P31749 | S83 | ATRGRGSSVGGGSRR | - |
| ASK1 | Q99683 | Y718 | IPERDSRYSQPLHEE | - |
| JAK2 | O60674 | Y798 | IPERDSRYSQPLHEE | |
| JAK1 | P23458 | Y798 | IPERDSRYSQPLHEE | |
| ASK1 | Q99683 | T813 | GDNVLINTYSGVLKI | |
| ASK1 | Q99683 | Y814 | DNVLINTYSGVLKIS | |
| ASK1 | Q99683 | T838 | GINPCTETFTGTLQY | + |
| PDK1 | O15530 | T838 | GINPCTETFTGTLQY | + |
| ASK1 | Q99683 | T842 | CTETFTGTLQYMAPE | - |
| ASK1 | Q99683 | S966 | NEYLRSISLPVPVLV | - |
| ASK1 | Q99683 | S1029 | DENFEDHSAPPSPEE | - |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ASK1 (MAP3K5) | Q99683 | S1029 | DENFEDHSAPPSPEE | - |
| ASK1 (MAP3K5) | Q99683 | S966 | NEYLRSISLPVPVLV | - |
| ASK1 (MAP3K5) | Q99683 | T813 | GDNVLINTYSGVLKI | |
| ASK1 (MAP3K5) | Q99683 | T838 | GINPCTETFTGTLQY | + |
| ASK1 (MAP3K5) | Q99683 | T842 | CTETFTGTLQYMAPE | - |
| ASK1 (MAP3K5) | Q99683 | Y718 | IPERDSRYSQPLHEE | - |
| ASK1 (MAP3K5) | Q99683 | Y814 | DNVLINTYSGVLKIS | |
| Cip1 (p21, CDKN1A) | P38936 | S97 | GGRRPGTSPALLQGT | |
| MKK3 (MAP2K3, MEK3) | P46734 | S218 | ISGYLVDSVAKTMDA | + |
| MKK4 (MAP2K4, MEK4) | P45985 | S257 | ISGQLVDSIAKTRDA | + |
| MKK4 (MAP2K4, MEK4) | P45985 | T261 | LVDSIAKTRDAGCRP | + |
| MKK6 (MAP2K6, MEK6) | P52564 | S207 | ISGYLVDSVAKTIDA | + |
| MKK6 (MAP2K6, MEK6) | P52564 | T211 | LVDSVAKTIDAGCKP | + |
| MKK7 (MAP2K7, MEK7) | O14733 | S271 | ISGRLVDSKAKTRSA | + |
| MKK7 (MAP2K7, MEK7) | O14733 | T275 | LVDSKAKTRSAGCAA | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
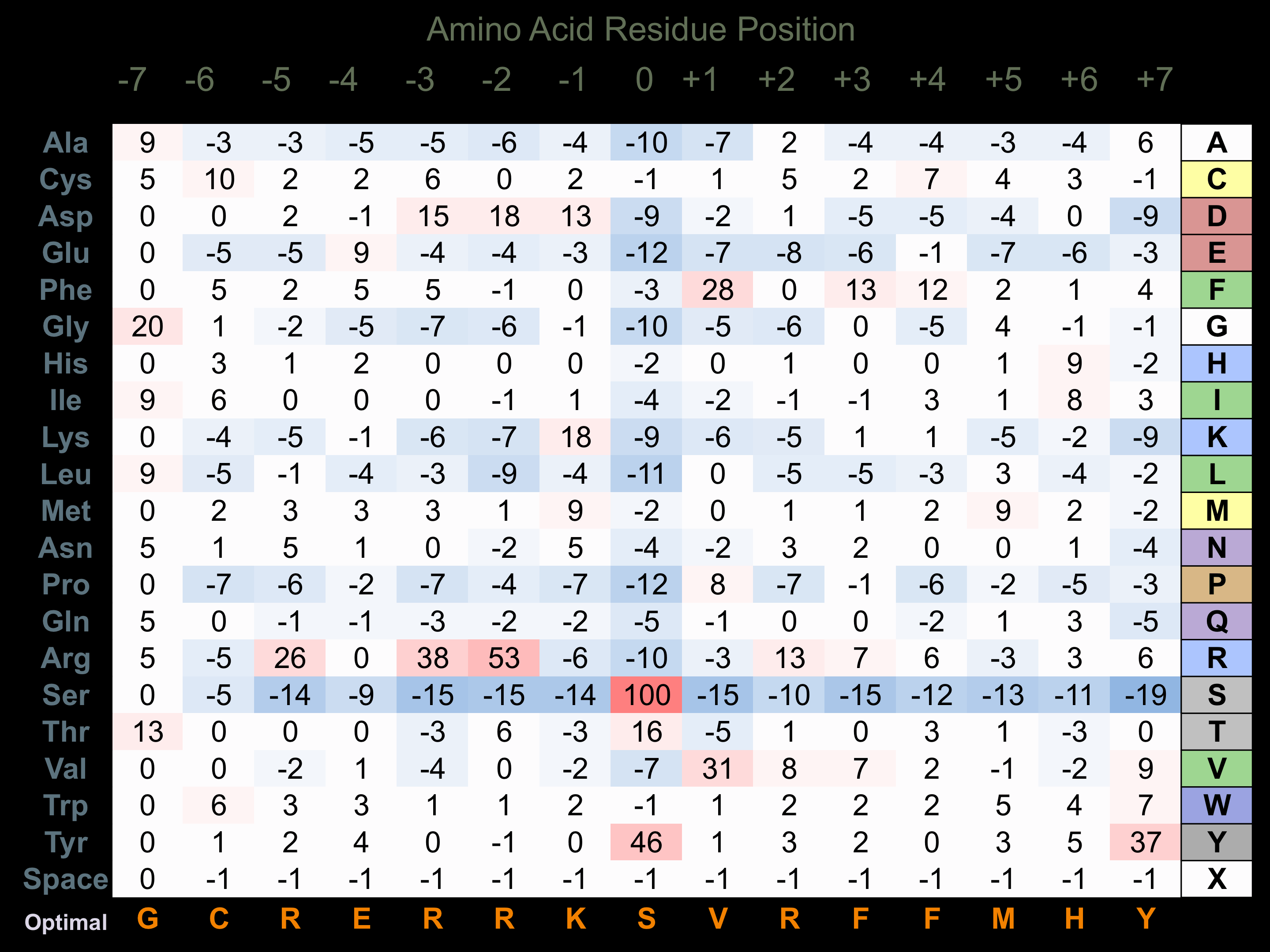
Matrix Type:
Experimentally derived from alignment of 18 known protein substrate phosphosites and 70 peptides phosphorylated by recombinant ASK1 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Melanomas
Comments:
ASK1 appears to be a tumour suppressor protein (TSP)The active form of the protein kinase normally acts to inhibit tumour cell proliferation. Mutations in the MAP3K5 gene have been observed in cancer patients, indicating a role for the protein in cancer pathology. For example, 24% of melanoma cell lines were found to contain mutations in the coding-regions of either MAP3K5 or the closely related kinase MAP3K9. Some of these mutations were found in the kinase catalytic domain, which were predicted to affect the catalytic activity of MAP3K5. In addition, loss of heterozygosity in MAP3K5 was observed in 85% of melanoma specimens, indicating that the previously observed mutations result in the loss-of-function in the protein. In vitro assays of MAP3K5 with an I780F substitution, the most commonly observed mutant form of the kinase, showed reduced phosphotransferase activity when compared to wild-type MAP3K5. It appears to be a tumour-suppressor protein with an important role in the prevention of cancer development.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +46, p<0.0007); Bladder carcinomas (%CFC= +95, p<0.0009); Cervical cancer stage 2B (%CFC= -64, p<0.081); Colon mucosal cell adenomas (%CFC= +53, p<0.0001); Oral squamous cell carcinomas (OSCC) (%CFC= +103, p<0.0001); Papillary thyroid carcinomas (PTC) (%CFC= +77, p<0.1); Pituitary adenomas (aldosterone-secreting) (%CFC= -57, p<0.0002); Skin melanomas - malignant (%CFC= -66, p<0.002); Skin squamous cell carcinomas (%CFC= +92, p<0.01); T-cell prolymphocytic leukemia (%CFC= -50, p<0.005); Uterine leiomyomas from fibroids (%CFC= -55, p<0.0002); and Vulvar intraepithelial neoplasia (%CFC= +75, p<0.0001). The COSMIC website notes an up-regulated expression score for ASK1 in diverse human cancers of 287, which is 0.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 40 for this protein kinase in human cancers was 0.7-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 25403 diverse cancer specimens. This rate is only -14 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.38 % in 805 skin cancers tested; 0.27 % in 602 endometrium cancers tested; 0.26 % in 1093 large intestine cancers tested; 0.15 % in 500 urinary tract cancers tested; 0.1 % in 1942 lung cancers tested; 0.06 % in 1466 breast cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: S53S (11).
Comments:
Only 4 deletions, 3 insertions, and 2 complex mutations are noted on the COSMIC website.