Nomenclature
Short Name:
MEK1
Full Name:
Dual specificity mitogen-activated protein kinase kinase 1
Alias:
- EC 2.7.12.2
- ERK activator kinase 1
- MAPK/ERK kinase 1
- MAPKK 1
- MAPKK1
- MKK1; PRKMK1
- Kinase MEK1
- MAP kinase kinase 1
- MAP2K1
- MAPK,ERK kinase 1
Classification
Type:
Dual specificity protein kinase
Group:
STE
Family:
STE7
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
43439
# Amino Acids:
393
# mRNA Isoforms:
2
mRNA Isoforms:
43,439 Da (393 AA; Q02750); 40,764 Da (367 AA; Q02750-2)
4D Structure:
Interacts with MORG1. Interacts with ARRB2 By similarity. Interacts with Yersinia yopJ.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 19 | 59 | Coiled-coil |
| 68 | 319 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K175, K362.
Serine phosphorylated:
S25, S72, S194, S212-, S218+, S222+, S231-, S298+, S385.
Threonine phosphorylated:
T23, T55, T226, T286-, T292-, T386+, T388+.
Tyrosine phosphorylated:
Y300.
Ubiquitinated:
K36, K57, K64, K97, K104, K168, K175, K192, K205, K353.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 62
62
981
16
1098
 20
20
318
12
222
 17
17
263
1
0
 100
100
1590
52
957
 66
66
1044
14
756
 17
17
268
45
174
 42
42
668
23
650
 34
34
535
27
649
 49
49
786
10
516
 17
17
273
61
152
 19
19
300
20
177
 41
41
649
140
670
 19
19
306
12
96
 13
13
209
12
104
 14
14
222
17
166
 18
18
283
9
85
 7
7
104
264
99
 15
15
234
10
160
 25
25
400
56
228
 49
49
774
56
656
 22
22
351
16
311
 17
17
276
18
263
 8
8
133
18
48
 14
14
227
10
156
 19
19
302
16
231
 61
61
962
30
767
 28
28
448
15
195
 18
18
287
10
171
 17
17
269
10
212
 4
4
57
14
37
 89
89
1415
18
695
 79
79
1251
21
1229
 71
71
1125
55
951
 77
77
1219
36
963
 28
28
440
22
426
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99
99
99.2
99 99.8
99.8
100
100 -
-
-
100 -
-
-
- -
-
-
99.5 -
-
-
- 99
99
99.5
99 99.2
99.2
99.8
99 -
-
-
- 73
73
77.9
- 81.7
81.7
88.9
96 91.9
91.9
96.2
93 33.3
33.3
51.4
89 -
-
-
- 62.4
62.4
77.8
68 -
-
-
- 52.4
52.4
69.2
54 64.8
64.8
75.7
- -
-
-
- -
-
-
- -
-
-
42 34.4
34.4
51.4
44 20.2
20.2
33.7
42 -
-
-
52
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | MAPK1 - P28482 |
| 2 | RAF1 - P04049 |
| 3 | MAPK3 - P27361 |
| 4 | BRAF - P15056 |
| 5 | ARAF - P10398 |
| 6 | MAPK8IP3 - Q9UPT6 |
| 7 | MAPKSP1 - Q9UHA4 |
| 8 | KSR1 - Q8IVT5 |
| 9 | MAPK14 - Q16539 |
| 10 | GRB10 - Q13322 |
| 11 | RPS6KA4 - O75676 |
| 12 | MAP3K8 - P41279 |
| 13 | HRAS - P01112 |
| 14 | CDK5 - Q00535 |
| 15 | EP300 - Q09472 |
Regulation
Activation:
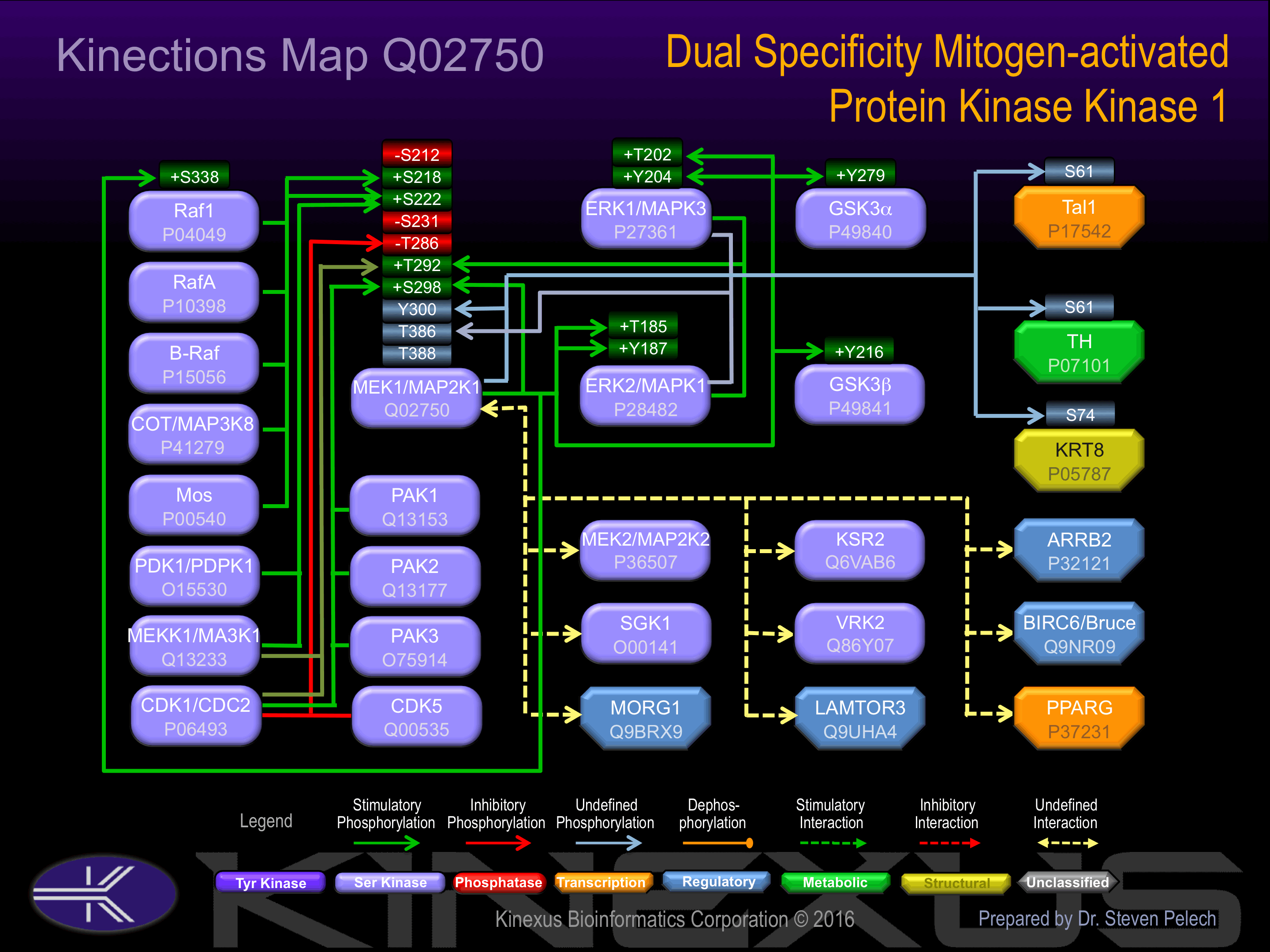
Phosphorylation at Ser-218 and Ser-222 increases phosphotransferase activity. Phosphorylation of Ser-298 increases phosphotransferse activity and association with Raf1 and ERK1.
Inhibition:
Phosphorylation of Ser-212 and Thr-286 inhibits phosphotransferase activity. Phosphorylation at Thr-292 increases molecular association with ERK2 and Raf1, but reduces interaction with ERK1. Phosphorylation of Thr-386 induces interaction with ERK2.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ERK1 | P27361 | T23 | DGSAVNGTSSAETNL | |
| ERK1 | P27361 | S25 | SAVNGTSSAETNLEA | |
| BRAF | P15056 | S218 | VSGQLIDSMANSFVG | + |
| MEKK1 | Q13233 | S218 | VSGQLIDSMANSFVG | + |
| COT | P41279 | S218 | VSGQLIDSMANSFVG | + |
| RAF1 | P04049 | S218 | VSGQLIDSMANSFVG | + |
| ARAF | P10398 | S218 | VSGQLIDSMANSFVG | + |
| MOS | P00540 | S218 | VSGQLIDSMANSFVG | + |
| BRAF | P15056 | S222 | LIDSMANSFVGTRSY | + |
| MEKK1 | Q13233 | S222 | LIDSMANSFVGTRSY | + |
| COT | P41279 | S222 | LIDSMANSFVGTRSY | + |
| PDK1 | O15530 | S222 | LIDSMANSFVGTRSY | + |
| RAF1 | P04049 | S222 | LIDSMANSFVGTRSY | + |
| ARAF | P10398 | S222 | LIDSMANSFVGTRSY | + |
| MOS | P00540 | S222 | LIDSMANSFVGTRSY | + |
| CDK5 | Q00535 | T286 | VEGDAAETPPRPRTP | - |
| CDK1 | P06493 | T286 | VEGDAAETPPRPRTP | - |
| ERK1 | P27361 | T292 | ETPPRPRTPGRPLSS | + |
| MEKK1 | Q13233 | T292 | ETPPRPRTPGRPLSS | + |
| CDK1 | P06493 | T292 | ETPPRPRTPGRPLSS | + |
| ERK2 | P28482 | T292 | ETPPRPRTPGRPLSS | - |
| MEK1 | Q02750 | S298 | RTPGRPLSSYGMDSR | + |
| PAK3 | O75914 | S298 | RTPGRPLSSYGMDSR | + |
| PAK1 | Q13153 | S298 | RTPGRPLSSYGMDSR | + |
| PAK2 | Q13177 | S298 | RTPGRPLSSYGMDSR | + |
| MEK1 | Q02750 | Y300 | PGRPLSSYGMDSRPP | |
| ERK1 | P27361 | T386 | IGLNQPSTPTHAAGV | ? |
| ERK2 | P28482 | T386 | IGLNQPSTPTHAAGV | ? |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ERK1 (MAPK3) | P27361 | T202 | HDHTGFLTEYVATRW | + |
| ERK1 (MAPK3) | P27361 | Y204 | HTGFLTEYVATRWYR | + |
| ERK2 (MAPK1) | P28482 | T185 | HDHTGFLTEYVATRW | + |
| ERK2 (MAPK1) | P28482 | Y187 | HTGFLTEYVATRWYR | + |
| GSK3a | P49840 | Y279 | RGEPNVSYICSRYYR | + |
| GSK3b | P49841 | Y216 | RGEPNVSYICSRYYR | + |
| KRT8 | P05787 | S74 | TVNQSLLSPLVLEVD | |
| MEK1 (MAP2K1) | Q02750 | S298 | RTPGRPLSSYGMDSR | + |
| MEK1 (MAP2K1) | Q02750 | Y300 | PGRPLSSYGMDSRPP | |
| Raf1 | P04049 | S338 | RPRGQRDSSYYWEIE | + |
| Tal1 | P17542 | S122 | DGRMVQLSPPALAAP | |
| TH | P07101 | S61 | SYTPTPRSPRFIGRR |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
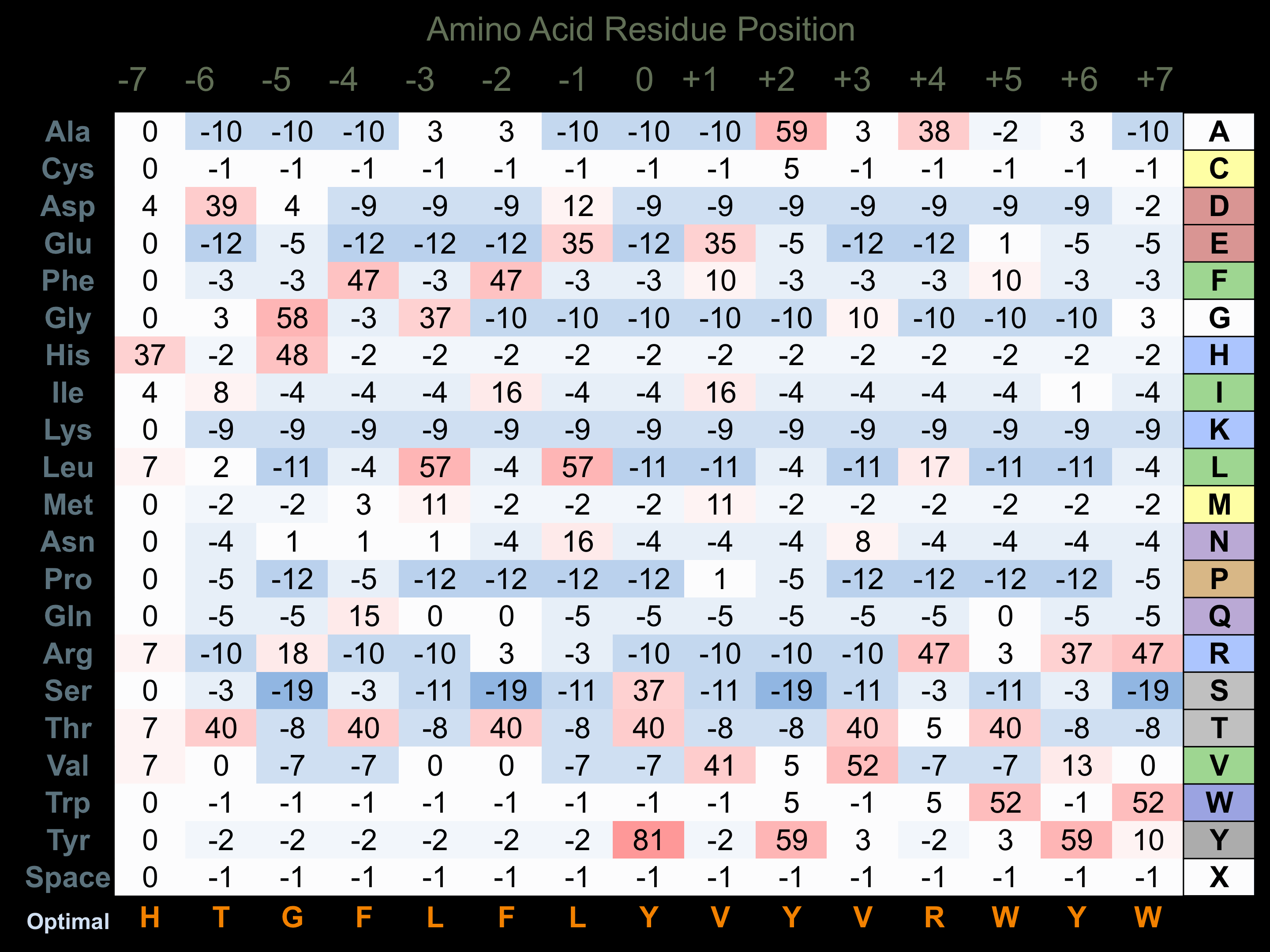
Matrix Type:
Experimentally derived from alignment of 22 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Developmental disorders
Specific Diseases (Non-cancerous):
Cardiofaciocutaneous syndrome; Cardiofaciocutaneous syndrome 3; Leopard syndrome; MAP2K1-related Noonan syndrome; MAP2K1-related cardiofaciocutaneous (CFC) syndrome
Comments:
Cardio-facio-cutaneous (CFC) syndrome is a disease characterized by abnormalities found in numerous parts of the body, particularly the heart, face, skin, and hair. Affected individuals display developmental delay and intellectual disability, ranging from moderate to severe. This disease is inherited in an autosomal dominant manner. Leopard syndrome is a genetic disease characterized by abnormalities in the heart, skin, inner ears, and genitalia. Most cases of this disease are inherited in an autosomal dominant manner. Noonan syndrome is a genetic disease characterized by the abnormal development of many parts of the body. Affected individuals display abnormal facial features, short stature, heart defects, and often delayed development. Mutations in the MEK1 gene have been observed in patients with CFC syndrome, including the F53S, Y130C, and G128V substitution mutations. MEK1 proteins containing the F53S and Y130C substitution mutations were shown to be unable to activate ERK/MAPK signalling unless they were phosphorylated by Raf at two serine phosphosites in the regulatory loop of the protein. Mutations in the MEK1 gene were observed in 5 out of 51 (9.8%) CFC syndrome patients, indicating a role for abnoraml MEK1 activity in the pathogenesis of the disease. In addition, ~60% of reported cases of Noonan syndrome are caused by mutations in proteins of the Ras-MAPK signalling pathway, specifically mutations that result in a gain-of-function of Raf protein activity. As MEK1 is an upstream activator of the Ras-MAPK signalling pathway, it has been implicated in the development of Noonan syndrome. Additionally, transgenic mice with overexpressed MEK1 protein specifically in the cardiomyocytes displayed substantial cardiac hypertrophy with the preservation of contractile function, indicating a link between MEK1 and cardiac disease, a common symptom of Noonan syndrome.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= -52, p<0.003); Bladder carcinomas (%CFC= +92, p<0.0002); Breast epithelial carcinomas (%CFC= -48, p<0.07); Cervical epithelial cancer (%CFC= +45, p<0.016); Prostate cancer - primary (%CFC= -76, p<0.0001); and Uterine fibroids (%CFC= +62, p<0.014). The COSMIC website notes an up-regulated expression score for MEK1 in diverse human cancers of 398, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 107 for this protein kinase in human cancers was 1.8-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.18 % in 31616 diverse cancer specimens. This rate is 2.3-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 1.22 % in 1379 skin cancers tested; 0.66 % in 1384 large intestine cancers tested; 0.45 % in 681 stomach cancers tested; 0.39 % in 2046 haematopoietic and lymphoid cancers tested; 0.14 % in 5486 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: P124S (29); K57N (16); C121S (10); E203K (9); D67N (9); F53L (8); P124L (5).
Comments:
Twelve deletions, no insertions and 2 complex mutations are noted on the COSMIC website.