Nomenclature
Short Name:
IKKb
Full Name:
Inhibitor of nuclear factor kappa B kinase beta subunit
Alias:
- EC 2.7.11.10
- IKKB
- IKK-beta
- NFKBIKB
- IKBKINASE
- I-kappa-B kinase 2
- I-kappa-B-kinase beta
- IkBKB
- IKK2
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
IKK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
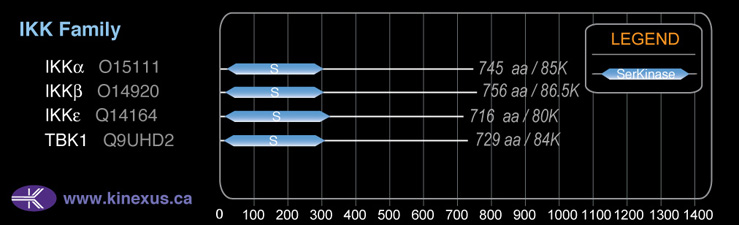
86,564
# Amino Acids:
756
# mRNA Isoforms:
4
mRNA Isoforms:
86,564 Da (756 AA; O14920); 85,945 Da (754 AA; O14920-2); 79,509 Da (697 AA; O14920-4); 29,236 Da (256 AA; O14920-3)
4D Structure:
Component of the I-kappa-B-kinase (IKK) core complex consisting of CHUK, IKBKB and IKBKG; probably four alpha/CHUK-beta/IKBKB dimers are associated with four gamma/IKBKG subunits. The IKK core complex seems to associate with regulatory or adapter proteins to form a IKK-signalosome holo-complex. Part of a complex composed of NCOA2, NCOA3, CHUK/IKKA, IKBKB, IKBKG and CREBBP. Part of a 70-90 kDa complex at least consisting of CHUK/IKKA, IKBKB, NFKBIA, RELA, IKBKAP and MAP3K14. Found in a membrane raft complex, at least composed of BCL10, CARD11, DPP4 and IKBKB. Interacts with SQSTM1 through PRKCZ or PRKCI. Forms an NGF-induced complex with IKBKB, PRKCI and TRAF6. May interact with MAVS/IPS1. Interacts with NALP2. Interacts with TICAM1. Interacts with Yersinia yopJ. Interacts with FAF1; the interaction disrupts the IKK complex formation. Interacts with ATM. Part of a ternary complex consisting of TANK, IKBKB and IKBKG. Interacts with NIBP; the interaction is direct. Interacts with ARRB1 and ARRB2. Interacts with TRIM21.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 15 | 312 | Pkinase |
| 458 | 479 | Leucine-zipper |
| 705 | 742 | IKK |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
T180, K659.
O-GlcNAcylated:
S733.
Serine phosphorylated:
S4, S6, S177+, S181+, S239, S246, S256, S257, S258, S267, S332, S335, S393, S402, S409, S411, S471, S474, S476, S489, S507, S550, S587, S600, S634, S670, S672, S675, S679, S682, S689, S692, S695, S697, S733-, S740-, S750-.
Threonine phosphorylated:
T23+, T180+, T324, T344, T368, T396, T399, T488, T559, T583, T610, T693, T735.
Tyrosine phosphorylated:
Y169, Y188-, Y199+, Y261, Y325, Y397, Y609.
Ubiquitinated:
K147, K163, K171, K418, K555, K703.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
988
42
1157
 5
5
49
20
43
 9
9
93
3
92
 44
44
433
128
992
 94
94
927
34
740
 7
7
73
119
102
 19
19
184
43
418
 86
86
851
52
1422
 49
49
486
24
384
 9
9
87
109
209
 9
9
91
36
179
 73
73
720
219
651
 9
9
93
36
83
 4
4
41
15
36
 20
20
196
26
340
 8
8
77
22
71
 27
27
271
377
355
 4
4
43
13
47
 10
10
94
111
94
 61
61
598
162
631
 18
18
178
24
328
 37
37
361
29
630
 6
6
62
6
55
 3
3
33
14
31
 47
47
463
24
793
 85
85
844
70
1241
 7
7
68
39
59
 5
5
48
14
45
 5
5
53
14
71
 5
5
50
42
59
 85
85
843
30
783
 67
67
660
51
649
 13
13
127
83
274
 87
87
857
78
717
 16
16
160
48
212
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 97.5
97.5
98.4
98.5 99.7
99.7
99.7
99.7 -
-
-
94 -
-
-
- 95
95
97.1
95 -
-
-
- 91.8
91.8
95.5
93 91.3
91.3
95.4
92 -
-
-
- 88.5
88.5
92.7
- 49.3
49.3
67.8
69.5 50.4
50.4
68.4
73 51.6
51.6
67.9
62 -
-
-
- 25
25
42.9
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | NFKBIA - P25963 |
| 2 | NFKB1 - P19838 |
| 3 | TNFRSF1A - P19438 |
| 4 | NFKBIB - Q15653 |
| 5 | RELA - Q04206 |
| 6 | PRKCQ - Q04759 |
| 7 | IRS1 - P35568 |
| 8 | CDC37 - Q16543 |
| 9 | TNFAIP3 - P21580 |
| 10 | RPS27A - P62988 |
| 11 | TSC1 - Q92574 |
| 12 | MAP3K11 - Q16584 |
| 13 | PRKDC - P78527 |
| 14 | PRKCB - P05771 |
| 15 | TRPC4AP - Q8TEL6 |
Regulation
Activation:
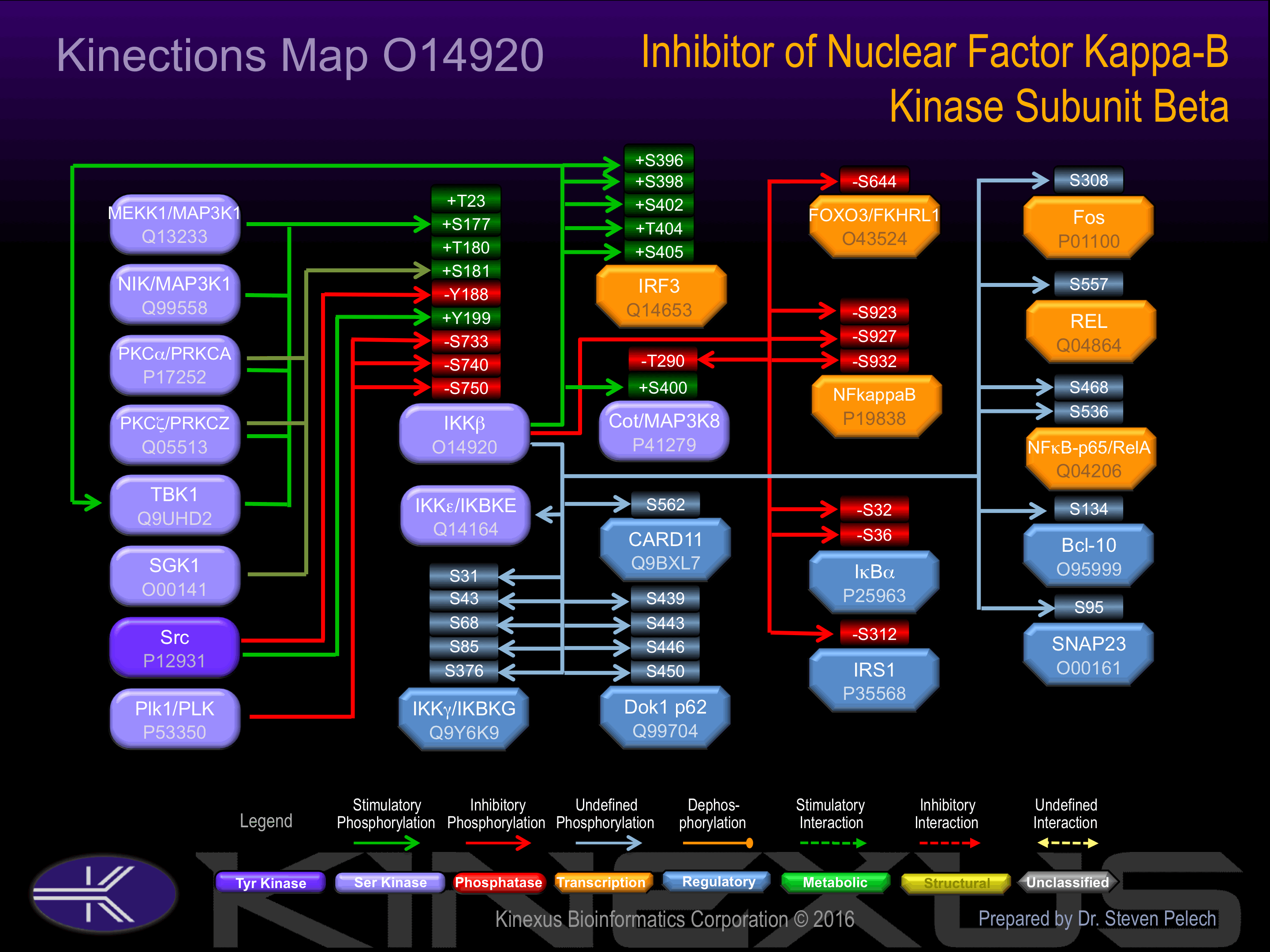
Activated by phosphorylation at Ser-177, Ser-181, Tyr-188 and Tyr-199 in the kinase activation loop.
Inhibition:
Inhibited by phosphorylation at Ser-733, Ser-740, Ser-750
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| PKCa | P17252 | S177 | AKELDQGSLCTSFVG | + |
| PKCz | Q05513 | S177 | AKELDQGSLCTSFVG | + |
| TBK1 | Q9UHD2 | S177 | AKELDQGSLCTSFVG | + |
| MEKK1 | Q13233 | S177 | AKELDQGSLCTSFVG | + |
| NIK | Q99558 | S177 | AKELDQGSLCTSFVG | + |
| PKCa | P17252 | S181 | DQGSLCTSFVGTLQY | + |
| PKCz | Q05513 | S181 | DQGSLCTSFVGTLQY | + |
| SGK | O00141 | S181 | DQGSLCTSFVGTLQY | + |
| SRC | P12931 | Y188 | SFVGTLQYLAPELLE | - |
| SRC | P12931 | Y199 | ELLEQQKYTVTVDYW | + |
| PLK1 | P53350 | S733 | TVREQDQSFTALDWS | - |
| PLK1 | P53350 | S740 | SFTALDWSWLQTEEE | - |
| PLK1 | P53350 | S750 | QTEEEEHSCLEQAS_ | - |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
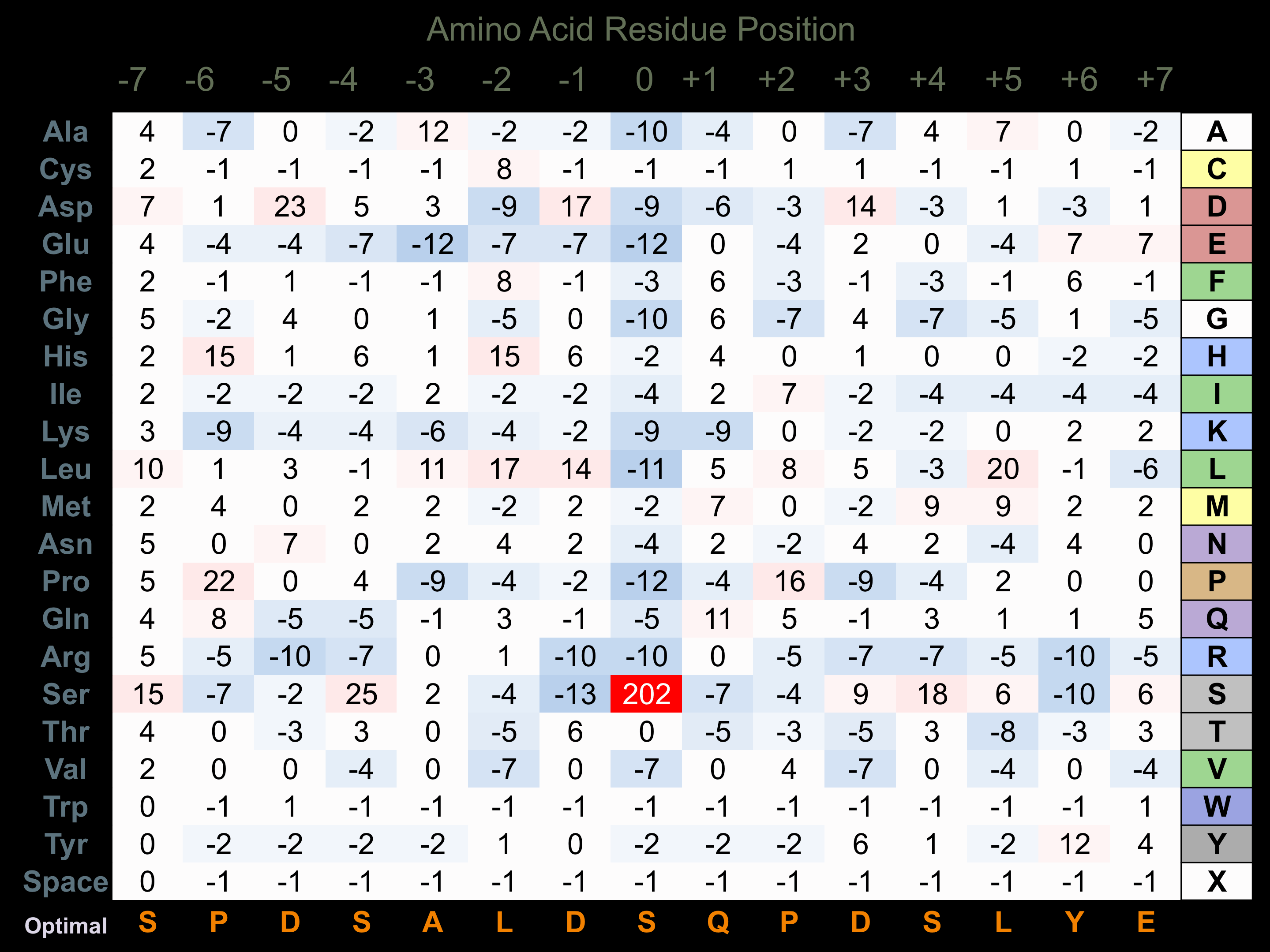
Matrix Type:
Experimentally derived from alignment of 61 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Endocrine disorders
Specific Diseases (Non-cancerous):
Insulin resistance; Immunodeficiency 15
Comments:
Insulin resistance is a disease characterized by a failure of cells to respond to normal levels of circulating insulin, resulting in hyperglycemia due to impaired glucose uptake. This causes an overproduction of insulin by the beta cells in the pancreas, which leads to hyperinsulinemia, and can contribute to the development of Type 2 diabetes or latent autoimmune diabetes in adults. immunodeficiency-15 (IMD15) is a immune disease characterized by the development in infants of severe bacterial, fungal, and viral infections, resulting in a failure to thrive. This disease is inherited in an autosomal recessive manner and results from defects in the differentiation and subsequent activation of immune cells (e.g. T and B cells). Mutations in the IKKb gene have been observed in patients with the IMD15 phenotype, including a 1-bp duplication mutation (c.1292dupG) in exon 13 of the gene resulting in a frameshift and premature truncation of the protein and loss of the alpha-helical dimerization domain, as well as a transition mutation resulting in an R272X mutation producing a truncated, non-functional protein correlated with the absence of memory B-cells and very low numbers of memory T-cells. In animal studies, mice lacking a functional IKKb protein specifically in myeloid cells displayed higher mortality when exposed to endotoxin, indicating a compromised immune system. In addition, the prolonged pharmacological inhibition of IKKb resulted in an increase in lipopolysaccharide-induced mortality in mice. Furthermore, inflammation has been proposed as a central component of several metabolic disorders, such as insulin resistance and type 2 diabetes. IKKb is a key coordinator of inflammatory responses through its role in the activation of NF-KB, indicating a potential mechanistic link. Interestingly, mice that lack IKKb expression in myeloid cells display the maintenance of insulin sensitivity and failure to develop insulin resistance even in response to a high fat diet, aging, or obesity.
Comments:
IKKb may be a tumour requiring protein (TRP), since it displays extremely low rates of mutation in human cancers.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +105, p<0.009); Brain glioblastomas (%CFC= -84, p<0.049); Brain oligodendrogliomas (%CFC= -87, p<0.044); Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= +90, p<0.088); Cervical cancer stage 1B (%CFC= +83, p<0.061); and Cervical cancer stage 2B (%CFC= +115, p<0.051). The COSMIC website notes an up-regulated expression score for IKKb in diverse human cancers of 666, which is 1.4-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 171 for this protein kinase in human cancers was 2.9-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.01 % in 25958 diverse cancer specimens. This rate is -90 % lower than the average rate of 0.075 % calculated for human protein kinases in general. Such a very low frequency of mutation in human cancers is consistent with this protein kinase playing a role as a tumour requiring protein (TRP).
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 2314 haematopoietic and lymphoid cancers tested; 0.01 % in 1683 breast cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: K171E (8); K171T )2).
Comments:
Only 5 deletions, no insertions or complex mutations are noted on the COSMIC website.