Nomenclature
Short Name:
CDK1
Full Name:
Cell division control protein 2 homolog
Alias:
- Cdc2
- CDC28
- Cyclin-dependent kinase 1
- EC 2.7.11.22
- EC 2.7.11.23
- P34 protein kinase; Kinase Cdc2; MPF;
- CDC28A
- CDC2A
- Cell division control protein 2
- Cell division cycle 2, G1 to S and G2 to M
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
CDK
SubFamily:
CDC2
Specific Links
Structure
Mol. Mass (Da):
34081
# Amino Acids:
297
# mRNA Isoforms:
2
mRNA Isoforms:
34,095 Da (297 AA; P06493); 27,503 Da (240 AA; P06493-2)
4D Structure:
Forms a stable but non-covalent complex with a regulatory subunit and with a cyclin. Interacts with DLGAP5. Isoform 2 is unable to complex with cyclin B1 and also fails to bind to the CDK inhibitor p21. Interacts with catalytically active CCNB1 and RALBP1 during mitosis to form an endocytotic complex during interphase.
3D Structure:
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 4 | 287 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K6, K9, K33, K34.
Serine phosphorylated:
S39, S46, S171, S178, S208, S233, S248, S265, S277.
Threonine phosphorylated:
T5, T14-, T141, T161+, T222, T240.
Tyrosine phosphorylated:
Y4-, Y15-, Y19-, Y77, Y160+, Y166-, Y270, Y286.
Ubiquitinated:
K9, K20, K24, K33, K34, K56, K89, K130, K139, K143, K201, K238, K243, K245, K254, K266, K274, K279, K295, K296.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-

 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 88.5
88.5
88.8
100 99.3
99.3
99.6
100 -
-
-
99 -
-
-
97 98.6
98.6
99.6
99 -
-
-
- 96.3
96.3
98.6
97 96.9
96.9
98.9
98 -
-
-
- 95.3
95.3
96.6
- 90.4
90.4
94.3
93 84.7
84.7
93.3
89 81.4
81.4
91.7
85 -
-
-
- 71.3
71.3
83.8
74 36.2
36.2
42.2
- 58.4
58.4
72.8
66 70.1
70.1
86.7
- -
-
-
- 63.3
63.3
78.7
- -
-
-
- 64.9
64.9
79.4
- 59
59
76.5
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CCNA2 - P20248 |
| 2 | CCNB2 - O95067 |
| 3 | CCNA1 - P78396 |
| 4 | TP53 - P04637 |
| 5 | CDC25C - P30307 |
| 6 | PKMYT1 - Q99640 |
| 7 | CCNE1 - P24864 |
| 8 | WEE1 - P30291 |
| 9 | LATS1 - O95835 |
| 10 | TSC1 - Q92574 |
| 11 | MYT1 - Q01538 |
| 12 | PBK - Q96KB5 |
| 13 | BIRC5 - O15392 |
| 14 | CDK7 - P50613 |
| 15 | GADD45G - O95257 |
Regulation
Activation:
Phosphorylation of Thr-161 increases phosphotransferase activity and interactions with cyclins CCNA2 and CCNB1. CCNA2 and CCNB1 both serve as activators of the catalytic activity of CDK1.
Inhibition:
Phosphorylation of Thr-14 or Tyr-15 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
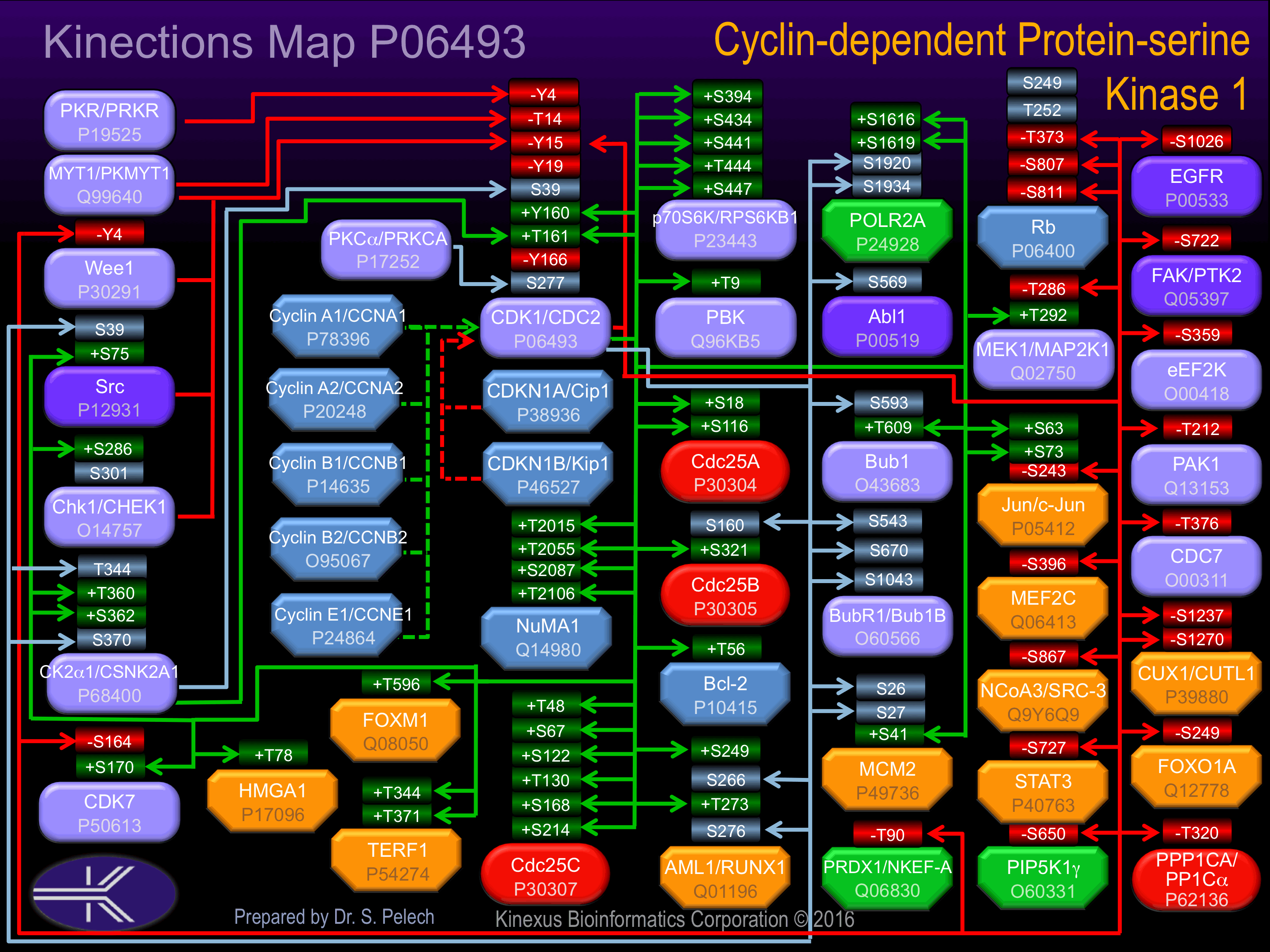
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| PKR | P19525 | Y4 | ____MEDYTKIEKIG | - |
| MYT1 | Q99640 | T14 | IEKIGEGTYGVVYKG | - |
| WEE1 | P30291 | Y15 | EKIGEGTYGVVYKGR | - |
| CHK1 | O14757 | Y15 | EKIGEGTYGVVYKGR | - |
| MYT1 | Q99640 | Y15 | EKIGEGTYGVVYKGR | - |
| CDK1 | P06493 | Y15 | EKIGEGTYGVVYKGR | - |
| SRC | P12931 | Y15 | EKIGEGTYGVVYKGR | - |
| CK2a1 | P68400 | S39 | MKKIRLESEEEGVPS | ? |
| CDK7 | P50613 | T161 | GIPIRVYTHEVVTLW | + |
| PKCa | P17252 | S277 | YDPAKRISGKMALNH |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| 4E-BP1 | Q13541 | T69 | RNSPVTKTPPRDLPT | |
| AAK1 | Q2M2I8 | T389 | LPIQPALTPRKRATV | |
| Abl1 | P00519 | S569 | LDHEPAVSPLLPRKE | |
| AHNAK | Q09666 | S5763 | EAEAEASSPKGKFSL | |
| AML1 (RUNX1) | Q01196 | S249 | DTRQIQPSPPWSYDQ | + |
| AML1 (RUNX1) | Q01196 | S266 | QYLGSIASPSVHPAT | |
| AML1 (RUNX1) | Q01196 | S276 | VHPATPISPGRASGM | |
| AML1 (RUNX1) | Q01196 | T273 | SPSVHPATPISPGRA | + |
| AML3 | Q01196 | S465 | MVPGGDRSPSRMLPP | |
| Amphiphysin | P49418 | S272 | EEPSPLPSPTASPNH | |
| Amphiphysin | P49418 | S276 | PLPSPTASPNHTLAP | |
| Amphiphysin | P49418 | S285 | NHTLAPASPAPARPR | |
| APC1 | Q9H1A4 | S355 | AALSRAHSPALGVHS | |
| APC1 | Q9H1A4 | S373 | VQRFNISSHNQSPKR | |
| APC1 | Q9H1A4 | S377 | NISSHNQSPKRHSIS | |
| APC1 | Q9H1A4 | S547 | PLSKLLGSLDEVVLL | |
| APC1 | Q9H1A4 | S555 | LDEVVLLSPVPELRD | |
| APC1 | Q9H1A4 | S688 | FDFEGSLSPVIAPKK | |
| APC1 | Q9H1A4 | T291 | KFSEQGGTPQNVATS | |
| APC1 | Q9H1A4 | T530 | NTMPRPSTPLDGVST | |
| APC1 | Q9H1A4 | T537 | TPLDGVSTPKPLSKL | |
| APC1 | Q9H1A4 | T701 | KKARPSETGSDDDWE | |
| APC4 | Q9UJX5 | S779 | KEEVLSESEAENQQA | |
| APP | P05067 | T743 | VEVDAAVTPEERHLS | |
| AR | P10275 | S81 | QQQQQETSPRQQQQQ | ? |
| ASE-1 | O15446 | S285 | PLEDTVLSPTKKRKR | |
| Bad | Q92934 | S91 | EGMGEEPSPFRGRSR | |
| BARD1 | Q99728 | S148 | NSIKMWFSPRSKKVR | |
| BARD1 | Q99728 | S251 | SQPSVISSPQINGEI | |
| BARD1 | Q99728 | S288 | PLAEQIESPDTKSRN | |
| BARD1 | Q99728 | T299 | KSRNEVVTPEKVCKN | |
| Bcl-2 | P10415 | T56 | FSSQPGHTPHPAASR | + |
| BIRC5 (Survivin) | O15392 | T34 | FLEGCACTPERMAEA | + |
| BLM | P54132 | S714 | PGVTVVISPLRSLIV | |
| BLM | P54132 | T766 | IIKLLYVTPEKICAS | |
| BORA | Q6PGQ7 | S252 | SSGQFSSSPIQASAK | |
| Bub1 | O43683 | S593 | CNKTLAPSPKSPGDF | |
| Bub1 | O43683 | T609 | SAAQLASTPFHKLPV | + |
| BubR1 (Bub1B) | O60566 | S1043 | WKVGKLTSPGALLFQ | |
| BubR1 (Bub1B) | O60566 | S543 | LSEKKNKSPPADPPR | |
| BubR1 (Bub1B) | O60566 | S670 | TLSIKKLSPIIEDSR | ? |
| Caldesmon | Q05682 | S511 | KHTENTFSRPGGRAS | |
| Caldesmon | Q05682 | S724 | EKGNVFSSPTAAGTP | |
| Caldesmon | Q05682 | S759 | KTPDGNKSPAPKPSD | |
| Caldesmon | Q05682 | S789 | QSVDKVTSPTKV___ | |
| Caldesmon | Q05682 | T638 | KKPFKCFTPKGSSLK | |
| Caldesmon | Q05682 | T730 | SSPTAAGTPNKETAG | |
| Caldesmon | Q05682 | T753 | INEWLTKTPDGNKSP | |
| Caldesmon iso3 | Q05682-3 | S524 | KTPDGNKSPAPKPSD | |
| Calnexin (CANX) | P27824 | S583 | EDEILNRSPRNRKPR | |
| CaRHSP1 | Q9Y2V2 | T23 | ASVGLLDTPRSRERS | |
| Caspase 9 | P55211 | T125 | PEVLRPETPRPVDIG | |
| Cdc16 | Q13042 | S112 | EKYLKDESGFKDPSS | |
| Cdc16 | Q13042 | S560 | KTLKNIISPPWDFRE | |
| Cdc16 | Q13042 | T581 | TAEETGLTPLETSRK | |
| Cdc23 | Q9UJX2 | T556 | QLRNQGETPTTEVPA | |
| Cdc23 | Q9UJX2 | T559 | NQGETPTTEVPAPFF | |
| Cdc25A | P30304 | S116 | PQKLLGCSPALKRSH | + |
| Cdc25A | P30304 | S18 | RRLLFACSPPPASQP | + |
| Cdc25B | P30305 | S160 | PVRLLGHSPVLRNIT | |
| Cdc25B | P30305 | S321 | KCQRLFRSPSMPCSV | + |
| Cdc25C | P30307 | S122 | DQHLMKCSPAQLLCS | + |
| Cdc25C | P30307 | S168 | SEMKYLGSPITTVPK | + |
| Cdc25C | P30307 | S214 | SRSGLYRSPSMPENL | + |
| Cdc25C | P30307 | T130 | PAQLLCSTPNGLDRG | + |
| Cdc25C | P30307 | T48 | VCPDVPRTPVGKFLG | + |
| Cdc25C | P30307 | T67 | LSILSGGTPKRCLDL | + |
| Cdc27 | P30260 | S222 | LNLESSNSKYSLNTD | |
| Cdc27 | P30260 | S312 | PPVIDVPSTGAPSKK | |
| Cdc27 | P30260 | S426 | TQPNINDSLEITKLD | |
| Cdc27 | P30260 | S434 | LEITKLDSSIISEGK | |
| Cdc27 | P30260 | S435 | EITKLDSSIISEGKI | |
| Cdc27 | P30260 | T205 | PETVLTETPQDTIEL | |
| Cdc27 | P30260 | T209 | LTETPQDTIELNRLN | |
| Cdc27 | P30260 | T289 | FGILPLETPSPGDGS | |
| Cdc27 | P30260 | T329 | ARIGQTGTKSVFSQS | |
| Cdc27 | P30260 | T343 | SGNSREVTPILAQTQ | |
| Cdc27 | P30260 | T430 | INDSLEITKLDSSII | |
| Cdc27 | P30260 | T446 | EGKISTITPQIQAFN | |
| CDC7 | O00311 | T376 | QVAPRAGTPGFRAPE | - |
| CDK1 (CDC2) | P06493 | Y15 | EKIGEGTYGVVYKGR | - |
| CDK7 | P50613 | S164 | GLAKSFGSPNRAYTH | - |
| CDK7 | P50613 | T170 | GSPNRAYTHQVVTRW | + |
| CEP55 | Q53EZ4 | S425 | NREKVAASPKSPTAA | |
| CEP55 | Q53EZ4 | S428 | KVAASPKSPTAALNE | |
| Chk1 (CHEK1) | O14757 | S286 | TSGGVSESPSGFSKH | + |
| Chk1 (CHEK1) | O14757 | S301 | IQSNLDFSPVNSASS | ? |
| CK2-B | Q5SRQ6 | S228 | QAASNFKSPVKTIR_ | |
| CK2a1 (CSNK2A1) | P68400 | S362 | ISSVPTPSPLGPLAG | + |
| CK2a1 (CSNK2A1) | P68400 | S370 | PLGPLAGSPVIAAAN | |
| CK2a1 (CSNK2A1) | P68400 | T344 | SSMPGGSTPVSSANM | |
| CK2a1 (CSNK2A1) | P68400 | T360 | SGISSVPTPSPLGPL | + |
| CKAP2 | Q8WWK9 | T623 | FKELKFLTPVRRSRR | |
| CREM | Q03060-2 | S147 | QGVVMAASPGSLHSP | |
| CREM | Q03060-2 | S153 | ASPGSLHSPQQLAEE | |
| CREM | Q03060-2 | S71 | EILSRRPSYRKILNE | |
| CREM | Q03060-2 | S94 | PKIEEERSEEEGTPP | |
| CTTN (Cortactin) | Q14247 | S405 | KTQTPPVSPAPQPTE | - |
| CUX1 (CUTL1) | P39880 | S1237 | TEYSQGASPQPQHQL | - |
| CUX1 (CUTL1) | P39880 | S1270 | YQQKPYPSPKTIEDL | - |
| Cyclin B1 | P14635 | S126 | PILVDTASPSPMETS | |
| Cyclin B1 | P14635 | S128 | LVDTASPSPMETSGC | |
| DDX3 | O00571 | T203 | LTRYTRPTPVQKHAI | |
| DDX3 | O00571 | T322 | GCHLLVATPGRLVDM | |
| Desmin | P17661 | S31 | PLGSPLSSPVFPRAG | |
| Desmin | P17661 | S6 | __SQSYSSSQRVSSY | |
| Desmin | P17661 | T78 | SRLGTTRTPSSYGAG | |
| Diaphanous 1 | O60610 | T747 | PVLPFGLTPKKLYKP | |
| DLG1 (SAP97) | Q12959 | S158 | FVSHSHISPIKPTEA | |
| DLG1 (SAP97) | Q12959 | S443 | FLGQTPASPARYSPV | |
| DLG7 | Q15398 | S618 | AGFFRVESPVKLFSG | |
| DLG7 | Q15398 | S839 | GGNLITFSPLQPGEF | |
| DLG7 | Q15398 | T329 | TYQVTPMTPRSANAF | |
| DLG7 | Q15398 | T639 | GPSQRLGTPKSVNKA | |
| DNCLI1 | Q9Y6G9 | S207 | PGEDFPASPQRRNTA | |
| DRP1 | O00429 | S616 | PIPIMPASPQKGHAV | |
| dUTPase | P33316 | S99 | GPETPAISPSKRARP | |
| dUTPase iso2 | P33316-2 | S11 | SEETPAISPSKRARP | |
| E2F1 | Q01094 | S332 | TDSATIVSPPPSSPP | |
| E2F1 | Q01094 | S337 | IVSPPPSSPPSSLTT | |
| ECT2 | Q9H8V3 | T342 | VSMLSLNTPNSNRKR | |
| ECT2 | Q9H8V3 | T413 | TKSSKSSTPVPSKQS | |
| ECT2 | Q9H8V3 | T815 | RAFSFSKTPKRALRR | |
| eEF1D iso1 | P29692 | S133 | APQTQHVSPMRQVEP | |
| eEF2K | O00418 | S359 | GTEEKCGSPRVRTLS | - |
| EFHD2 | Q96C19 | S74 | QGIGEPQSPSRRVFN | |
| EGFR | P00533 | S1026 | PQQGFFSSPSTSRTP | - |
| ELAVL1 | Q15717 | S202 | LLSQLYHSPARRFGG | |
| Ensconsin (MAP7) | Q14244 | S209 | SSATLLNSPDRARRL | |
| EPB41 iso2 | P11171-2 | S679 | DKRLSTHSPFRTLNI | |
| EPB41 iso2 | P11171-2 | T60 | LKASNGDTPTHEDLT | |
| Epsin 1 | Q9Y6I3 | S357 | FSDPWGGSPAKPSTN | |
| ERCC6L | Q2NKX8 | T1063 | VKQFDASTPKNDISP | |
| FAK (PTK2) | Q05397 | S722 | PSRPGYPSPRSSEGF | - |
| FANCG | O15287 | S387 | PRFSPPPSPPGPCMP | |
| FEN1 | P39748 | S187 | MDCLTFGSPVLMRHL | |
| FLNA | P21333 | S1436 | GGHQVPGSPFKVPVH | |
| FLNA | P21333 | S1533 | GDEEVPRSPFKVKVL | |
| FLNA | P21333 | S1630 | GGDEIPFSPYRVRAV | |
| FOXM1 | Q08050 | T611 | ETLPISSTPSKSVLP | + |
| FOXO1A | Q12778 | S249 | EGGKSGKSPRRRAAS | - |
| GAPVD1 | Q14C86 | S977 | APHSSSSSPSKDSSR | |
| GIGYF2 | Q6Y7W6 | S30 | SITSPPLSPALPKYK | |
| GMPS | P49915 | T318 | PISDEDRTPRKRISK | |
| GRP58 | P30101 | S456 | GFPTIYFSPANKKLN | |
| H1E | P10412 | T17 | APAPAEKTPVKKKAR | |
| Hamartin | Q92574 | S584 | ETSIFTPSPCKIPPP | |
| Hamartin | Q92574 | T1047 | SSSSELSTPEKPPHQ | |
| Hamartin | Q92574 | T417 | SLPQATVTPPRKEER | |
| HMGA1 | P17096 | T78 | KTRKTTTTPGRKPRG | + |
| HMGA1 iso2 | P17096-2 | T41 | KEPSEVPTPKRPRGR | |
| HMGCS1 | Q01581 | S495 | IATEHIPSPAKKVPR | |
| hnRNP K | P61978 | S216 | ILDLISESPIKGRAQ | + |
| HR6A | P49459 | S120 | LDEPNPNSPANSQAA | |
| INCENP | Q9NQS7 | T412 | DTEIANSTPNPKPAA | |
| INCENP | Q9NQS7 | T59 | EPELMPKTPSQKNRR | |
| IP3R1 (IP3 receptor) | Q14643 | S421 | VMLKIGTSPVKEDKE | |
| IP3R1 (IP3 receptor) | Q14643 | T800 | RDPQEQVTPVKYARL | |
| IREB2 | P48200 | S157 | LQKAGKLSPLKVQPK | |
| IRS2 | Q9Y4H2 | S391 | SVAGSPLSPGPVRAP | |
| Jun (c-Jun) | P05412 | S243 | PGETPPLSPIDMESQ | - |
| Jun (c-Jun) | P05412 | S63 | KNSDLLTSPDVGLLK | + |
| Jun (c-Jun) | P05412 | S73 | VGLLKLASPELERLI | + |
| JunB | P17275 | S189 | YTNLSSYSPASASSG | |
| JunB | P17275 | T153 | LHKMNHVTPPNVSLG | |
| KIF11 | P52732 | T926 | LDIPTGTTPQRKSYL | |
| KIF22 | Q14807 | S427 | ASASQKLSPLQKLSS | |
| KIF22 | Q14807 | T463 | QGAPLLSTPKRERMV | |
| KRT8 | P05787 | S432 | SAYGGLTSPGLSYSL | |
| Lamin A,C | P02545 | S22 | QASSTPLSPTRITRL | |
| Lamin A,C | P02545 | S390 | EEERLRLSPSPTSQR | |
| Lamin A,C | P02545 | S392 | ERLRLSPSPTSQRSR | |
| Lamin A/C | P02545 | T19 | SGAQASSTPLSPTRI | |
| Lamin B1 | P20700 | S22 | EEERLRLSPSPTSQR | |
| Lamin B1 | P20700 | S390 | EEERLKLSPSPSSRV | |
| Lamin B1 | P20700 | S391 | EEERLKLSPSPSSRV | |
| Lamin B1 | P20700 | S392 | GGPTTPLSPTRLSRL | |
| Lamin B1 | P20700 | T19 | SRAGGPTTPLSPTRL | |
| LAP2A | P42166 | S423 | FQETEFLSPPRKVPR | |
| LATS1 | O95835 | S613 | EKKQITTSPITVRKN | |
| LATS1 | O95835 | T490 | ATTVTAITPAPIQQP | |
| LBR | Q14739 | S71 | KGGSTSSSPSRRRGS | |
| LIG1 | P18858 | S76 | EEEDEALSPAKGQKP | |
| LIG3 | P49916 | S210 | TTTGQVTSPVKGASF | |
| LIG3 | P49916 | S913 | EKLATKSSPVKVGEK | |
| LIG3 | P49916 | T191 | LSSKAAGTPKKKAVV | |
| MAP4 | P27816 | S696 | PNKELPPSPEKKTKP | |
| MAP4 | P27816 | S787 | KAPEKRASPSKPASA | - |
| MAP4 | P27816 | T892 | VPSRVKATPMPSRPS | |
| MAP4 | P27816 | T901 | MPSRPSTTPFIDKKP | |
| MAP4 | P27816 | T917 | SAKPSSTTPRLSRLA | |
| MASTL | Q96GX5 | T741 | DDGRILGTPDYLAPE | - |
| MCL1 | Q07820 | S64 | IGGSAGASPPSTLTP | |
| MCM2 | P49736 | S26 | RGNDPLTSSPGRSSR | |
| MCM2 | P49736 | S27 | GNDPLTSSPGRSSRR | ? |
| MCM2 | P49736 | S41 | RTDALTSSPGRDLPP | + |
| MCM3 | P25205 | S112 | SFGSKHVSPRTLTSC | |
| MCM3 | P25205 | S611 | SSDTARTSPVTARTL | |
| MCM3 | P25205 | T722 | EEMPQVHTPKTADSQ | |
| MCM4 | P33991 | S3 | _____MSSPASTPSR | |
| MCM4 | P33991 | S32 | RSEDARSSPSQRRRG | |
| MCM4 | P33991 | S88 | PLDFDVSSPLTYGTP | |
| MCM4 | P33991 | T110 | PRSGVRGTPVRQRPD | |
| MCM4 | P33991 | T19 | GSRRGRATPAQTPRS | |
| MCM4 | P33991 | T7 | _MSSPASTPSRRSSR | |
| MDM4 | O15151 | S96 | SFSVKDPSPLYDMLR | |
| MEF2C | Q06413 | S396 | NIKSEPVSPPRDRTT | - |
| MEK1 (MAP2K1) | Q02750 | T286 | VEGDAAETPPRPRTP | - |
| MEK1 (MAP2K1) | Q02750 | T292 | ETPPRPRTPGRPLSS | + |
| MKI67 | P46013 | T761 | GIAEMFKTPVKEQPQ | |
| MKI67IP (NIFK) | Q9BYG3 | T238 | QGPTPVCTPTFLERR | ? |
| MyoD | P15172 | S200 | SGDSDASSPRSNCSD | |
| MyoD | P15172 | S5 | MELLSPPLRDVD___ | |
| MYPT1 | O14974 | S409 | SGQATPTSPIKKFPT | |
| MYPT1 | O14974 | S432 | EEERKDESPATWRLG | |
| MYPT1 | O14974 | S473 | GVTRSASSPRLSSSL | |
| MYPT1 | O14974 | S601 | TKITTGSSSAGTQSS | |
| MYST2 | O95251 | T85 | TRSQQQPTPVTPKKY | |
| MYST2 | O95251 | T88 | QQQPTPVTPKKYPLR | |
| NAGK | Q9UJ70 | S76 | RSLGLSLSGGDQEDA | |
| NCAPG | Q9BPX3 | T308 | LNALFSITPLSELVG | ? |
| NCAPG | Q9BPX3 | T332 | LIPVETLTPEIALYW | ? |
| NCAPG | Q9BPX3 | T931 | KNKEVYMTPLRGVKA | |
| NCL | P19338 | T121 | PGKALVATPGKKGAA | |
| NCOA3 (SRC-3) | Q9Y6Q9 | S728 | VVKQEQLSPKKKENN | ? |
| NCoA3/SRC-3 | Q9Y6Q9 | S867 | SPVSVGSSPPVKNIS | - |
| NDEL1 | Q9GZM8 | S242 | IPNGFGTSPLTPSAR | |
| NDEL1 | Q9GZM8 | T219 | ASLSLPATPVGKGTE | |
| NDEL1 | Q9GZM8 | T245 | GFGTSPLTPSARISA | |
| NM23 (NME1) | P15531 | S120 | GRNIIHGSDSVESAE | |
| NM23 (NME1) | P15531 | T94 | GRVMLGETNPADSKP | |
| NME2 | P22392 | T94 | GRVMLGETNPADSKP | |
| NME2P1 | O60361 | T79 | GRVMLGETNPADSKP | |
| Nogo | Q9NQC3 | S15 | PLVSSSDSPPRPQPA | |
| NOLC1 | Q14978 | T607 | AKKIKLQTPNTFPKR | |
| NOLC1 | Q14978 | T610 | IKLQTPNTFPKRKKG | |
| NPM1 | P06748 | S70 | EAMNYEGSPIKVTLA | |
| NPM1 | P06748 | T199 | VKKSIRDTPAKNAQK | |
| NPM1 | P06748 | T219 | KDSKPSSTPRSKGQE | |
| NPM1 | P06748 | T234 | SFKKQEKTPKTPKGP | |
| NPM1 | P06748 | T237 | KQEKTPKTPKGPSSV | |
| NSBP1 | P82970 | T31 | SAMLVPVTPEVKPKR | |
| NSFL1C | Q9UNZ2 | S140 | AVERVTKSPGETSKP | |
| NUCKS1 | Q9H1E3 | S181 | LKATVTPSPVKGKGK | |
| NuMA1 | Q14980 | S2087 | TRSGTRRSPRIATTT | + |
| NuMA1 | Q14980 | T2015 | SCFPRPMTPRDRHEG | + |
| NuMA1 | Q14980 | T2055 | MAFSILNTPKKLGNS | + |
| NuMA1 | Q14980 | T2106 | TAAAIGATPRAKGKA | + |
| NUP133 | Q8WUM0 | T28 | AGLGPGSTPRTASRK | |
| NUP210 | Q8TEM1 | T1844 | LAVPAALTPRASPGH | |
| NUP210 | Q8TEM1 | V1839 | CTPRDLAvPAALTPR | |
| NUP35 | Q8NFH5 | S138 | GTGQSMFSPASIGQP | |
| NUP35 | Q8NFH5 | S259 | SDRCALSSPSLAFTP | |
| NUP35 | Q8NFH5 | T106 | SSPGLGSTPLTSRRQ | |
| NUP35 | Q8NFH5 | T273 | PPIKTLGTPTQPGST | |
| ODF2 | Q5BJF6 | S820 | YSTFLTSSPIRSRSP | |
| OGFR | Q9NZT2 | S378 | EDRPEPLSPKESKKR | |
| ORC6L | Q9Y5N6 | T195 | REPGDVATPPRKRKK | |
| p53 | P04637 | S315 | LPNNTSSSPQPKKKP | + |
| p70S6K (RPS6KB1) | P23443 | S394 | TRQTPVDSPDDSTLS | + |
| p70S6K (RPS6KB1) | P23443 | S434 | SFEPKIRSPRRFIGS | + |
| p70S6K (RPS6KB1) | P23443 | S441 | SPRRFIGSPRTPVSP | + |
| p70S6K (RPS6KB1) | P23443 | S447 | GSPRTPVSPVKFSPG | + |
| p70S6K (RPS6KB1) | P23443 | T444 | RFIGSPRTPVSPVKF | + |
| p73 | O15350 | T86 | AASASPYTPEHAASV | |
| PAICS | P22234 | S27 | EVYELLDSPGKVLLQ | |
| PAK1 | Q13153 | T212 | VIEPLPVTPTRDVAT | - |
| Palladin | Q8WX93 | S641 | PTPAVLLSPTKEPPP | |
| Palladin | Q8WX93 | S766 | SSLPSPMSPTPRQFG | |
| PBK | Q96KB5 | T9 | EGISNFKTPSKLSEK | + |
| PDS5B | Q9NTI5 | T1370 | AIESTQSTPQKGRGR | |
| PIK3C2A | O00443 | S259 | KVSNLQVSPKSEDIS | |
| PIK4CB | Q9UBF8 | S266 | PAPDTGLSPSKRTHQ | |
| PIP5KG | O60331 | S650 | DERSWVYSPLHYSAQ | - |
| PITPNM1 | O00562 | S382 | DFIDAFASPVEAEGT | ? |
| PITPNM1 | O00562 | T287 | SAASNTGTPDGPEAP | |
| PKN2 (PRK2) | Q16513 | S535 | VNHSGTFSPQAPVPT | |
| plectin 1 | P30427 | T4402 | GGLIEPDTPGRVPLD | |
| POLR2A | P24928 | S1616 | TPQSPSYSPTSPSYS | + |
| POLR2A | P24928 | S1619 | SPSYSPTSPSYSPTS | + |
| POLR2A | P24928 | S1920 | SPTYSPTSPKYSPTS | |
| POLR2A | P24928 | S1934 | SPTYSPTSPKGSTYS | |
| POM121 | Q96HA1 | T203 | HVYPSLPTPLLRPSR | |
| PPP1CA | P62138 | T320 | NPGGRPITPPRNSAK | - |
| PPP1R1A | Q13522 | S67 | LKSTLAMSPRQRKKM | |
| PRC1 | O43663 | T481 | RRGLAPNTPGKARKL | |
| PRDX1 (NKEF-A) | Q06830 | T90 | CHLAWVNTPKKQGGL | - |
| PTP1B | P18031 | S386 | LRGAQAASPAKGEPS | |
| PTPN2 | P17706 | S304 | LSPAFDHSPNKIMTE | |
| PYCR2 | Q96C36 | S304 | VSTLTPSSPGKLLTR | |
| Rab4A | P20338 | S199 | AALRQLRSPRRAQAP | |
| Rab5B | P61020 | S123 | KELQRQASPSIVIAL | |
| Rad9 | Q99638 | S277 | SHSQDLGSPERHQPV | |
| Rad9 | Q99638 | S328 | VLPSISLSPGPQPPK | ? |
| Rad9 | Q99638 | S336 | PGPQPPKSPGPHSEE | |
| Rad9 | Q99638 | T292 | PQLQAHSTPHPDDFA | |
| Rad9 | Q99638 | T355 | EPSTVPGTPPPKKFR | |
| RanBP2 | P49792 | S21 | SVQGSTPSPRQKSMK | |
| RanBP2 | P49792 | S2251 | HASPLASSPVRKNLF | |
| RanBP2L2 | P0DJD0 | S1275 | HASPLASSPVRKNLF | |
| RanGAP1 | P46060 | S442 | STFLAFPSPEKLLRL | |
| RanGAP1 | P46060 | T409 | GQGEKSATPSRKILD | |
| Rb | P06400 | S249 | AVIPINGSPRTPRRG | |
| Rb | P06400 | S807 | PGGNIYISPLKSPYK | - |
| Rb | P06400 | S811 | IYISPLKSPYKISEG | - |
| Rb | P06400 | T252 | PINGSPRTPRRGQNR | |
| Rb | P06400 | T373 | VNVIPPHTPVRTVMN | - |
| RCC1 | P18754 | S11 | KRIAKRRSPPADAIP | |
| RCC1 | P18754 | S387 | GQDEDAWSPVEMMGK | |
| RCC1 | P18754 | T274 | SNYHQLGTPGTESCF | |
| REPS2 | Q8NFH8 | S463 | RPRSRSYSSTSIEEA | |
| RFC1 | P35251 | S518 | VQGKRKISPSKKESE | |
| RFC1 | P35251 | T506 | KESKLERTPQKNVQG | |
| RGPD3 | A6NKT7 | S1276 | HASPLASSPVRKNLF | |
| RGPD5 | Q99666 | S1275 | HASPLASSPVRKNLF | |
| RPA2 | P15927 | S29 | QSPGGFGSPAPSQAE | |
| RRM2 | P31350 | S20 | DPQQLQLSPLKGLSL | |
| SAMHD1 | Q9Y3Z3 | T592 | DVIAPLITPQKKEWN | |
| Separase | Q14674 | S1126 | IAPSTNSSPVLKTKP | |
| SIRT1 | Q96EB6 | S540 | HVSEDSSSPERTSPP | |
| SIRT1 | Q96EB6 | T530 | YLSELPPTPLHVSED | |
| SIRT2 | Q8IXJ6 | S368 | PNPSTSASPKKSPPP | |
| SPT5 | O00267 | S666 | VGGFAPMSPRISSPM | |
| Src | P12931 | S75 | NSSDTVTSPQRAGPL | + |
| Src | P12931 | T37 | GAFPASQTPSKPASA | |
| SREBP1 | P36956 | S439 | AGSPFQSSPLSLGSR | |
| SRRM2 | Q9UQ35 | T1413 | VLDAVPRTPSRERSS | |
| SRRM2 | Q9UQ35 | T866 | QANEQSVTPQRRSCF | |
| SSR1 | P43307 | S268 | LNQINKASPRRLPRK | |
| STAT3 | P40763 | S727 | NTIDLPMSPRTLDSL | - |
| STI1 | P31948 | T198 | DEEEEIATPPPPPPP | |
| STI1 | P31948 | T332 | KSLAEHRTPDVLKKC | |
| STMN1 | P16949 | S25 | QAFELILSPRSKESV | |
| STMN1 | P16949 | S38 | SVPEFPLSPPKKKDL | |
| STMN2 | Q93045 | S73 | EAPRTLASPKKKDLS | |
| SYN3 | O14994 | S470 | PQGQQPLSPQSGSPQ | |
| TCOF1 | Q13428 | S156 | ANLLSGKSPRKSAEP | |
| TCOF1 | Q13428 | S583 | LQAKPTSSPAKGPPQ | |
| TCOF1 | Q13428 | T983 | AQAQAASTPRKARAS | |
| TERF1 | P54274 | T344 | KKERRVGTPQSTKKK | + |
| TERF1 | P54274 | T371 | VSKSQPVTPEKHRAR | + |
| TFPT | P0C1Z6 | S180 | PAPPEPGSPAPGEGP | |
| Tip60 | Q92993 | S90 | LPGSRPGSPEREVPA | |
| TK1 | P04183 | S13 | LPTVLPGSPSKTRGQ | |
| TMEM48 | Q9BTX1 | T414 | PEETAFQTPKSSQMP | |
| TMEM48 | Q9BTX1 | T415 | PEETAFQTPKPSQMP | |
| TMPO | P42167 | T160 | GTESRSSTPLPTISS | |
| TMPO | P42167 | T279 | GPVISESTPIAETIM | |
| TOP1 | P11387 | S112 | EKENGFSSPPQIKDE | |
| TOP1 | P11387 | S394 | SKDAKVPSPPPGHKW | |
| TOP2A | P11388 | S1213 | QMAEVLPSPRGQRVI | |
| TOP2A | P11388 | S1247 | KNENTEGSPQEDGVE | |
| TOP2A | P11388 | S1354 | DFVPSDASPPKTKTS | |
| TOP2A | P11388 | S1361 | SPPKTKTSPKLSNKE | |
| TOP2A | P11388 | S1393 | GSVPLSSSPPATHFP | |
| TOP2B | Q02880 | S1424 | DKDEYTFSPGKSKAT | |
| TOR1AIP1 | Q5JTV8 | S143 | EQPPLQPSPVMTRRG | |
| TPP1 | O14773 | T201 | VGLHLGVTPSVIRKR | |
| TPX2 | Q9ULW0 | S738 | QPLTVPVSPKFSTRF | |
| TTDN1 | Q8TAP9 | S104 | SQQQFGYSPGQQQTH | |
| TTDN1 | Q8TAP9 | S93 | YPGSYSRSPAGSQQQ | |
| TTDN1 | Q8TAP9 | T120 | QGSPRTSTPFGSGRV | |
| TUBA3C | Q13748 | T62 | RRPESFTTPEGPKPR | |
| TUBB | P07437 | S172 | NTFSVVPSPKVSDTV | |
| U2AF2 | P26368 | S79 | EHGGLIRSPRHEKKK | |
| UBA1 | P22314 | S4 | ____MSSSPLSKKRR | |
| UBAP2L | Q14157 | S454 | AAPPPPSSPLPSKST | |
| UGDH | O60701 | T461 | TIGFQIETIGKKVSS | |
| UNG | P13051 | S64 | EPGTPPSSPLSAEQL | |
| USP14 | P54578 | T235 | SSSASAATPSKKKSL | |
| Vimentin | P08670 | S55 | TSRSLYASSPGGVYA | |
| Vimentin | P08670 | S56 | SRSLYASSPGGVYAT | |
| Wee1 | P30291 | S123 | EEGFGSSSPVKSPAA | - |
| WIBG | Q9BRP8 | T72 | PEATAPVTPSRPEGG | |
| ZC3H11A | O75152 | T321 | PETNIDKTPKKAQVS | |
| ZC3HC1 (NIPA) | Q86WB0 | S395 | PGLEVPSSPLRKAKR | - |
| ZMYM3 | Q14202 | T826 | PPPPPPATPRKNKAA |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
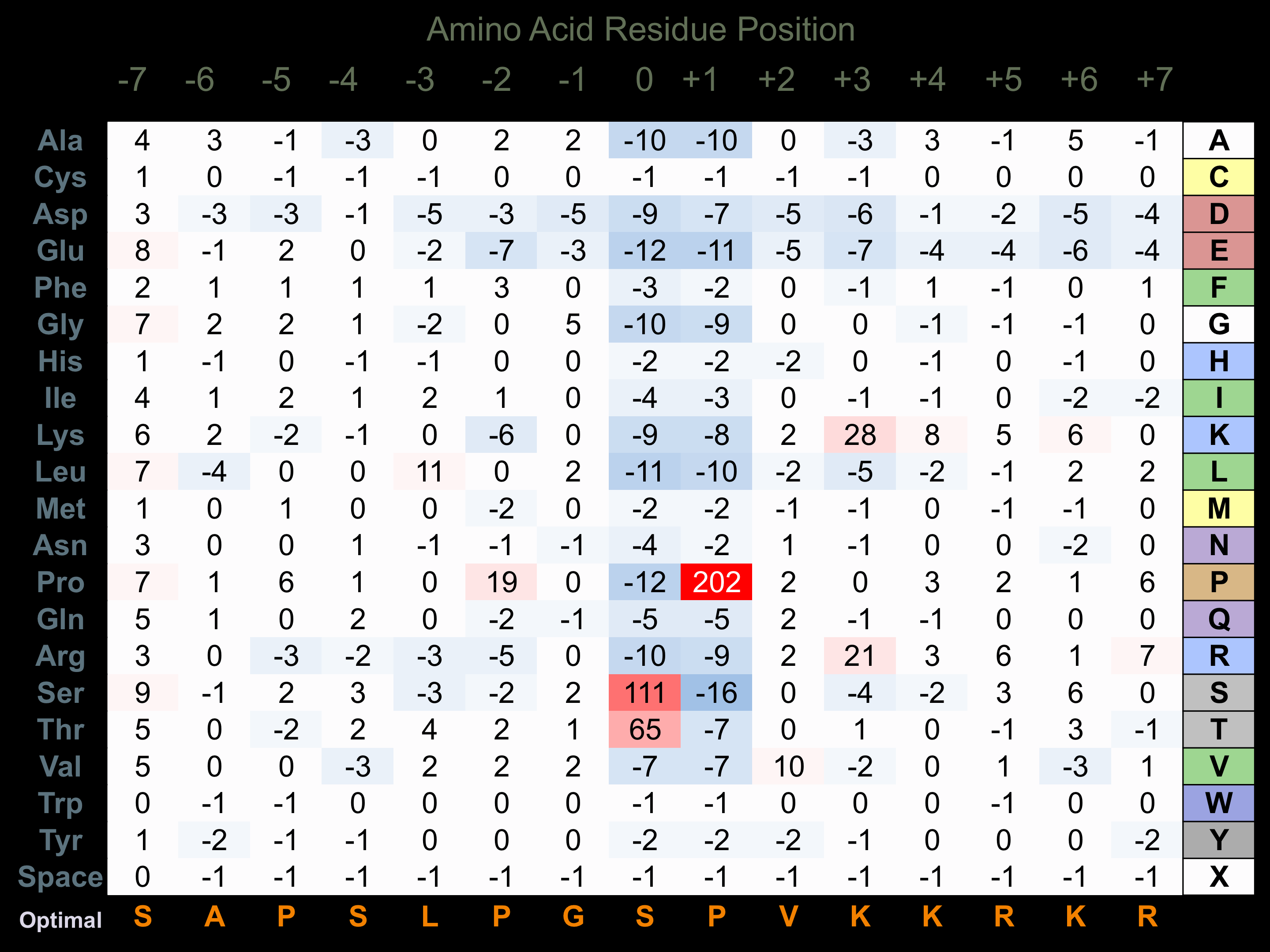
Matrix Type:
Experimentally derived from alignment of 538 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Thyroid lymphomas; Male germ cell tumours
Comments:
CDK1 appears to be a tumour requiring protein (TRP). Gain-of-function mutations in the CDK1 gene have been linked to several forms of cancer, indicating an oncogenic role for the CDK1 protein. Cells transformed with the oncogene MYC undergo apoptosis when treated with small-molecule CDK1 inhibitors. CDK1-inhibitor treatment of MYC-dependent lymphoma and hepatoblastoma cells isolated from mice resulted in decreased tumour growth and prolonged cell survival. In addition, the elevated expression of CDK1 has been reported as a diagnostic marker for cancer progression in esophageal adenocarcinoma, potentially reflecting the role of the CDK1 protein in tumorigenesis. In addition, loss of the normal regulatory mechanisms of CDK1 activity in LZTS1-null mice was observed to predispose the mice to both spontaneous and carcinogen-induced cancer development, indicating that aberrant CDK1 expression functions to promote cancer formation. Furthermore, CDK1 expression can be used as a prognostic indicator for early breast cancer. For example, breast cancer tumours with high expression of CDK1 are correlated with a significantly lower 5-year patient survival rate (66. 9%) than tumours that have low levels of CDK1 expression (84. 2%).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 23713 diverse cancer specimens. This rate is only -32 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 2.07 % in 1184 large intestine cancers tested; 1.31 % in 589 stomach cancers tested; 1.04 % in 805 skin cancers tested; 0.95 % in 602 endometrium cancers tested; 0.78 % in 904 ovary cancers tested; 0.74 % in 500 urinary tract cancers tested; 0.6 % in 1942 lung cancers tested; 0.41 % in 816 prostate cancers tested; 0.34 % in 1467 breast cancers tested; 0.27 % in 1226 kidney cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R257Q (3).
Comments:
No deletions, insertions or complex mutations are noted on the COSMIC website.