Nomenclature
Short Name:
PKCd
Full Name:
Protein kinase C, delta type
Alias:
- EC 2.7.11.13
- PKC-delta
- PRKCD
- Protein kinase C, delta
- Kinase PKC-delta
- KPCD
- MAY 1
- MGC49908
- NPKC-delta
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
PKC
SubFamily:
Delta
Specific Links
Structure
Mol. Mass (Da):
77505
# Amino Acids:
676
# mRNA Isoforms:
2
mRNA Isoforms:
80,953 Da (707 AA; Q05655-2); 77,505 Da (676 AA; Q05655)
4D Structure:
Interacts with PDK1, RAD9A, CDCP1 and MUC1
1D Structure:
Subfamily Alignment
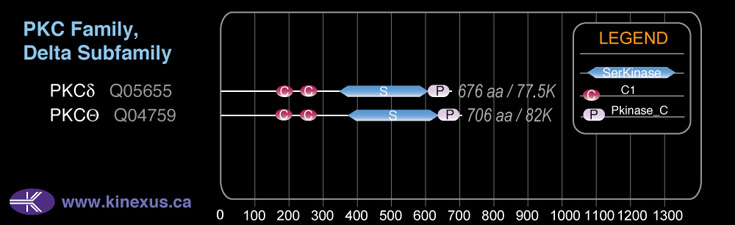
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S130, S299, S302, S304, S306, S307, S331, S341, S342-, S359-, S503+, S506+, S631, S645+, S647, S654, S658, S664.
Threonine phosphorylated:
T43, T50, T141+, T218, T295+, T333, T350, T451, T507+, T511-, T594, T605, T609.
Tyrosine phosphorylated:
Y52-, Y64-, Y155-, Y187-, Y313+, Y334, Y374, Y448, Y453, Y471+, Y514-, Y525+, Y567-, Y630, Y646.
Ubiquitinated:
K184, K222, K271, K319, K336, K362, K369, K496, K613, K641.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 67
67
975
16
1118
 14
14
201
15
97
 12
12
180
18
118
 15
15
214
78
379
 38
38
547
17
650
 46
46
666
53
1177
 38
38
554
26
649
 100
100
1454
52
2714
 36
36
527
13
602
 10
10
147
79
120
 4
4
63
37
63
 40
40
576
148
587
 13
13
186
29
143
 2
2
34
15
23
 4
4
58
18
41
 13
13
192
9
169
 15
15
223
207
1283
 9
9
137
30
201
 2
2
22
69
20
 41
41
596
59
658
 7
7
109
34
143
 12
12
170
36
116
 25
25
361
27
199
 8
8
123
30
126
 6
6
94
34
65
 59
59
857
58
1566
 12
12
177
35
115
 14
14
197
30
167
 5
5
79
30
70
 2
2
24
14
14
 65
65
947
30
634
 32
32
460
21
554
 15
15
214
55
333
 72
72
1051
31
613
 88
88
1276
22
2283
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 62.8
62.8
76.8
100 58.5
58.5
59
99 -
-
-
88 -
-
-
86 63.6
63.6
77.6
92 -
-
-
- 90.4
90.4
95.4
91 89.5
89.5
95.3
90 -
-
-
- 63.7
63.7
78
- 82.4
82.4
90
86 81.4
81.4
89.6
83 33.3
33.3
50.9
79 -
-
-
- 46.8
46.8
62
57.5 45.3
45.3
61.4
- 47.2
47.2
64.8
51 49.1
49.1
63.6
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | INSR - P06213 |
| 2 | STAT3 - P40763 |
| 3 | IRS1 - P35568 |
| 4 | YWHAZ - P63104 |
| 5 | PEBP1 - P30086 |
| 6 | EGFR - P00533 |
| 7 | PRKDC - P78527 |
| 8 | LMNB2 - Q03252 |
| 9 | PLSCR3 - Q9NRY6 |
| 10 | ADRB2 - P07550 |
| 11 | ADCY7 - P51828 |
| 12 | PLD2 - O14939 |
| 13 | MEP1B - Q16820 |
| 14 | MAPK3 - P27361 |
| 15 | ESRRA - P11474 |
Regulation
Activation:
Phosphorylation at Tyr-313, Tyr-514, Tyr-525, Ser-645 increases phosphotransferase activity. Phosphorylation of Thr-507 increases phosphotransferase activity and it can induce degradation of PKC-delta.
Inhibition:
Phosphorylation of Tyr-52, Tyr-64, Tyr-187, and Tyr-567 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| LYN | P07948 | Y52 | VQKKPTMYPEWKSTF | - |
| SRC | P12931 | Y64 | STFDAHIYEGRVIQI | - |
| PKCd | Q05655 | T141 | EDEAKFPTMNRRGAI | + |
| SRC | P12931 | Y155 | IKQAKIHYIKNHEFI | - |
| LYN | P07948 | Y155 | IKQAKIHYIKNHEFI | - |
| SRC | P12931 | Y187 | WGLNKQGYKCRQCNA | - |
| FYN | P06241 | Y187 | WGLNKQGYKCRQCNA | - |
| PKCd | Q05655 | T218 | TAANSRDTIFQKERF | |
| PKCd | Q05655 | T295 | AEALNQVTQRASRRS | + |
| PKCd | Q05655 | S302 | TQRASRRSDSASSEP | |
| PKCd | Q05655 | S304 | RASRRSDSASSEPVG | |
| PDGFRA | P16234 | Y313 | SSEPVGIYQGFEKKT | + |
| YES | P07947 | Y313 | SSEPVGIYQGFEKKT | + |
| SRC | P12931 | Y313 | SSEPVGIYQGFEKKT | + |
| FYN | P06241 | Y313 | SSEPVGIYQGFEKKT | + |
| PKCd | Q05655 | Y313 | SSEPVGIYQGFEKKT | + |
| EGFR | P00533 | Y313 | SSEPVGIYQGFEKKT | + |
| LYN | P07948 | Y313 | SSEPVGIYQGFEKKT | + |
| LCK | P06239 | Y313 | SSEPVGIYQGFEKKT | + |
| PDGFRA | P16234 | Y334 | MQDNSGTYGKIWEGS | ? |
| YES | P07947 | Y334 | MQDNSGTYGKIWEGS | ? |
| SRC | P12931 | Y334 | MQDNSGTYGKIWEGS | ? |
| FYN | P06241 | Y334 | MQDNSGTYGKIWEGS | ? |
| PKCd | Q05655 | Y334 | MQDNSGTYGKIWEGS | ? |
| EGFR | P00533 | Y334 | MQDNSGTYGKIWEGS | ? |
| LYN | P07948 | Y334 | MQDNSGTYGKIWEGS | ? |
| LCK | P06239 | Y334 | MQDNSGTYGKIWEGS | ? |
| SRC | P12931 | Y374 | ELKGRGEYSAIKALK | |
| PKCd | Q05655 | S503 | KENIFGESRASTFCG | + |
| PDK1 | O15530 | T507 | FGESRASTFCGTPDY | + |
| SRC | P12931 | T507 | FGESRASTFCGTPDY | + |
| PKCe | Q02156 | T507 | FGESRASTFCGTPDY | + |
| LCK | P06239 | Y514 | TFCGTPDYIAPEILQ | - |
| SRC | P12931 | Y567 | IRVDTPHYPRWITKE | - |
| LYN | P07948 | Y567 | IRVDTPHYPRWITKE | - |
| SRC | P12931 | Y630 | KVKSPRDYSNFDQEF | |
| PKCd | Q05655 | S645 | LNEKARLSYSDKNLI | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| 14-3-3 beta (YWHAB) | P31946 | S59 | NVVGARRSSWRVISS | |
| 14-3-3 beta (YWHAB) | P31946 | S60 | VVGARRSSWRVISSI | |
| 14-3-3 eta | Q04917 | S58 | VVGARRSSWRVISSI | |
| 14-3-3 zeta (YWHAZ) | P63104 | S58 | VVGARRSSWRVVSSI | |
| ADAM17 | P78536 | T735 | KPFPAPQTPGRLQPA | |
| AML3 | Q13950 | S240 | KLDDSKPSLFSDRLS | |
| BARK1 (GRK2, ADRBK1) | P25098 | S29 | ATPAARASKKILLPE | + |
| C5aR | P21730 | S334 | SVVRESKSFTRSTVD | |
| ChAT | P28329 | S558 | VPTYESASIRRFQEG | |
| ChAT | P28329 | S594 | HKAAVPASEKLLLLK | |
| DAB2 | P98082 | S24 | QAAPKAPSKKEKKKG | |
| DAP3 | P51398 | S185 | FDQPLEASTWLKNFK | |
| DAP3 | P51398 | S280 | TLKREDKSPIAPEEL | |
| DAP3 | P51398 | S31 | MGTQARQSIAAHLDN | |
| DAP3 | P51398 | T186 | DQPLEASTWLKNFKT | |
| DAP3 | P51398 | T237 | ITRVRNATDAVGIVL | |
| DBI | P07108 | T41 | ATVGDINTERPGMLD | |
| DBI | P07108 | T64 | AWNELKGTSKEDAMK | |
| eEF1A-1 | P68104 | T432 | AVRDMRQTVAVGVIK | |
| ELAVL1 | Q15717 | S221 | QAQRFRFSPMGVDHM | |
| ELAVL1 | Q15717 | S318 | GDKILQVSFKTNKSH | |
| eNOS | P29474 | S1177 | TSRIRTQSFSLQERQ | + |
| EP300 | Q09472 | S89 | SELLRSGSSPNLNMG | - |
| FLI1 | Q01543 | T312 | TNGEFKMTDPDEVAR | + |
| GAP43 | P17677 | S41 | AATKIQASFRGHITR | |
| Gg(12) | Q9UBI6 | S2 | _______SSKTASTN | |
| gp130 | P40189 | T890 | GQVERFETVGMEAAT | |
| GSK3a | P49840 | S21 | SGRARTSSFAEPGGG | - |
| GSK3b | P49841 | S9 | SGRPRTTSFAESCKP | - |
| H2B | P33778 | S15 | APAPKKGSKKAITKA | |
| H3.1 | P68431 | T45 | PHRYRPGTVALREIR | |
| HMGA1 | P17096 | S44 | PGTALVGSQKEPSEV | |
| HMGA1 | P17096 | S64 | PRGRPKGSKNKGAAK | |
| hnRNP K | P61978 | S302 | GRGGRGGSRARNLPL | |
| HSP27 | P04792 | S15 | FSLLRGPSWDPFRDW | ? |
| HSP27 | P04792 | S82 | RALSRQLSSGVSEIR | ? |
| IRS1 | P35568 | S24 | GYLRKPKSMHKRFFV | |
| IRS1 | P35568 | S307 | TRRSRTESITATSPA | - |
| IRS1 | P35568 | S323 | MVGGKPGSFRVRASS | |
| IRS1 | P35568 | S441 | SPCDFRSSFRSVTPD | |
| IRS1 | P35568 | S574 | RLPGHRHSAFVPTRS | |
| IRS1 | P35568 | S616 | DDGYMPMSPGVAPVP | - |
| ITGB2 | P05107 | S745 | FEKEKLKSQWNNDNP | |
| ITGB2 | P05107 | T758 | NPLFKSATTTVMNPK | |
| JNK1 (MAPK8) | P45983 | S129 | ELDHERMSYLLYQML | + |
| KRT8 | P05787 | S74 | TVNQSLLSPLVLEVD | |
| LIMK2 | P53671 | S283 | EGTLRRRSLRRSNSI | ? |
| M-CK | P06732 | S128 | LDPNYVLSSRVRTGR | |
| Met | P08581 | S985 | PHLDRLVSARSVSPT | - |
| MUC1 | P15941 | T1224 | RYVPPSSTDRSPYEK | |
| MYBPC3 | Q14896 | S304 | SLLKKSSSFRTPRDS | |
| p47phox | P14598 | S303 | RGAPPRRSSIRNAHS | + |
| p47phox | P14598 | S304 | GAPPRRSSIRNAHSI | + |
| p47phox | P14598 | S315 | AHSIHQRSRKRLSQD | ? |
| p47phox | P14598 | S320 | QRSRKRLSQDAYRRN | ? |
| p47phox | P14598 | S328 | QDAYRRNSVRFLQQR | + |
| p47phox | P14598 | S359 | EERQTQRSKPQPAVP | + |
| p47phox | P14598 | S370 | PAVPPRPSADLILNR | + |
| p47phox | P14598 | S379 | DLILNRCSESTKRKL | ? |
| p53 | P04637 | S15 | PSVEPPLSQETFSDL | + |
| p53 | P04637 | S46 | AMDDLMLSPDDIEQW | + |
| p70S6Kb (RPS6KB2) | Q9UBS0 | S473 | PPSGTKKSKRGRGRP | + |
| p73 iso2 | O15350 | S289 | GQVLGRRSFEGRICA | |
| PA2G4 | Q9UQ80 | S360 | ELKALLQSSASRKTQ | ? |
| PEBP1 | P30086 | S153 | RGKFKVASFRKKYEL | |
| PKCd (PRKCD) | Q05655 | S302 | TQRASRRSDSASSEP | |
| PKCd (PRKCD) | Q05655 | S304 | RASRRSDSASSEPVG | |
| PKCd (PRKCD) | Q05655 | S503 | KENIFGESRASTFCG | + |
| PKCd (PRKCD) | Q05655 | S645 | LNEKARLSYSDKNLI | + |
| PKCd (PRKCD) | Q05655 | T141 | EDEAKFPTMNRRGAI | + |
| PKCd (PRKCD) | Q05655 | T218 | TAANSRDTIFQKERF | |
| PKCd (PRKCD) | Q05655 | T295 | AEALNQVTQRASRRS | + |
| PKCd (PRKCD) | Q05655 | Y313 | SSEPVGIYQGFEKKT | + |
| PKCd (PRKCD) | Q05655 | Y334 | MQDNSGTYGKIWEGS | ? |
| PKD1 (PRKCM) | Q15139 | S738 | ARIIGEKSFRRSVVG | + |
| PKD1 (PRKCM) | Q15139 | S742 | GEKSFRRSVVGTPAY | + |
| PLCB3 | Q01970 | S1105 | LDRKRHNSISEAKMR | - |
| PLSCR1 | O15162 | T161 | CGPSRPFTLRIIDNM | |
| PLSCR3 | Q9NRY6 | T21 | PPPPYPVTPGYPEPA | |
| PPARA | Q07869 | S110 | RICGDKASGYHYGVH | + |
| PPARA | Q07869 | S142 | VYDKCDRSCKIQKKN | + |
| PPARA | Q07869 | S179 | RFGRMPRSEKAKLKA | ? |
| PPP1R14A (CPI 17) | Q96A00 | T38 | QKRHARVTVKYDRRE | - |
| PPP1R14B | Q96C90 | T57 | VRRQGKVTVKYDRKE | - |
| PTPN22 | Q9Y2R2 | S35 | FLKLKRQSTKYKADK | - |
| RasGRP3 | Q8IV61 | T133 | YDWMRRVTQRKKVSK | |
| RGS9 | O75916-3 | S478 | MSKLDRRSQLKKELP | |
| S6 | P62753 | S235 | IAKRRRLSSLRASTS | + |
| S6 | P62753 | S236 | AKRRRLSSLRASTSK | + |
| Shc1 | P29353 | S139 | EEWTRHGSFVNKPTR | |
| SMPD1 | P17405 | S508 | DGNYSRSSHVVLDHE | |
| Src | P12931 | S12 | KSKPKDASQRRRSLE | + |
| STAT1 | P42224 | S727 | TDNLLPMSPEEFDEV | - |
| STAT3 | P40763 | S727 | NTIDLPMSPRALDSL | - |
| syndecan-4 | P31431 | S179 | MKKKDEGSYDLGKKP | |
| TBL1XR1 | Q9BZK7 | S123 | AAASQQGSAKNGENT | |
| TIF1b | Q13263 | S473 | SGVkRSRSGEGEVSG | + |
| TNNI3 | P19429 | S22 | PAPIRRRSSNYRAYA | |
| TNNI3 | P19429 | S23 | APIRRRSSNYRAYAT | |
| TNNI3 | P19429 | S41 | AKKKSKISASRKLQL | |
| TNNI3 | P19429 | S43 | KKSKISASRKLQLKT | |
| TNNI3 | P19429 | S76 | GEKGRALSTRCQPLE | |
| TNNI3 | P19429 | T142 | RGKFKRPTLRRVRIS | |
| TNNT2 | P45379 | S207 | QAQTERKSGKRQTER | |
| TNNT2 | P45379 | T203 | YIQKQAQTERKSGKR | |
| TNNT2 | P45379 | T212 | RKSGKRQTEREKKKK | |
| TNNT2 | P45379 | T293 | TRGKAKVTGRWK___ | |
| TRPC6 | Q9Y210 | S449 | KFVAHAASFTIFLGL | - |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
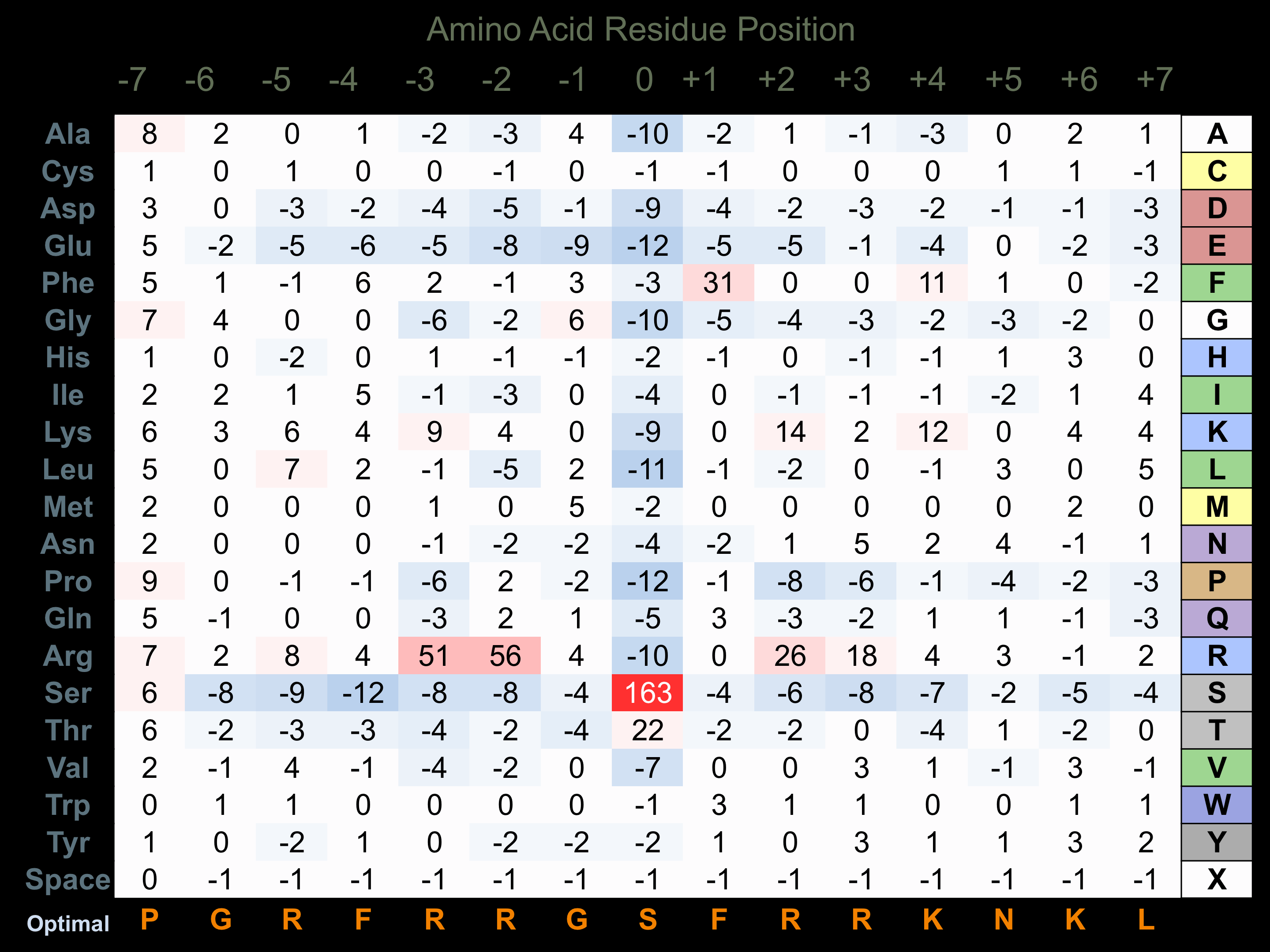
Matrix Type:
Experimentally derived from alignment of 167 known protein substrate phosphosites and 34 peptides phosphorylated by recombinant PKCd in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, immune disorders
Specific Diseases (Non-cancerous):
Immunodeficiency, common variable, 9; Common variable Immune deficiency; Autoimmune lymphoproliferative syndrome, Type 1b; Systemic lupus erythematosus 16
Comments:
Reduced PKCd expression or activity is associated with immunodeficiency, common variable, 9, which is a primary immunodeficiency and results in B-cell deficiency and severe autoimmunity. Mutations are also associated with autoimmune lymphoproliferative syndrome type III, indicating this gene has a crucial role in regulating B-cell tolerance and preventing self-reactivity in humans.
Specific Cancer Types:
ACTH-secreting pituitary adenomas
Comments:
PKCd may be a tumour suppressor protein (TSP). PKCd negatively regulates cell proliferation in colon cell lines. It is also required for ErbB2-driven mammary gland tumorigenesis, and negatively correlates with prognosis in human breast cancer.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +134, p<0.0005); Brain glioblastomas (%CFC= +4697, p<0.0001); Brain oligodendrogliomas (%CFC= +10153, p<0.0001); Breast epithelial cell carcinomas (%CFC= +74, p<0.01); Cervical cancer (%CFC= +61, p<0.011); Skin fibrosarcomas (%CFC= -48); and Skin melanomas - malignant (%CFC= +569, p<0.0001). The COSMIC website notes an up-regulated expression score for PKCd in diverse human cancers of 336, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 119 for this protein kinase in human cancers was 2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25347 diverse cancer specimens. This rate is only -4 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.51 % in 1093 large intestine cancers tested; 0.35 % in 805 skin cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R6L (4); R6H (2).
Comments:
Only 5 deletions (all at D434fs*2), 2 insertions (1 at D434fs*13), and no complex mutations are noted on the COSMIC website. About 32% of the point mutations are silent and do not change the amino acid sequence of the protein kinase.

