Nomenclature
Short Name:
RAF1
Full Name:
RAF proto-oncogene serine-threonine-protein kinase
Alias:
- CRAF
- Murine leukemia viral (V-raf-1) oncogene homologue 1
- Protein kinase raf 1
- Raf proto-oncogene serine/threonine-protein kinase
- RAF1
- V-raf-1 murine leukemia viral oncogene 1
- C-RAF
- C-Raf
- EC 2.7.11.1
- Kinase Raf1
Classification
Type:
Protein-serine/threonine kinase
Group:
TKL
Family:
RAF
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
73,052
# Amino Acids:
648
# mRNA Isoforms:
2
mRNA Isoforms:
75,395 Da (668 AA; P04049-2); 73,052 Da (648 AA; P04049)
4D Structure:
Forms a multiprotein complex with Ras (M-Ras/MRAS), SHOC2 and protein phosphatase 1 (PPP1CA, PPP1CB and PPP1CC). Interacts with Ras proteins; the interaction is antagonized by RIN1. Weakly interacts with RIT1 By similarity. Interacts with STK3; the interaction inhibits its pro-apoptotic activity. Interacts (when phosphorylated at Ser-259) with YWHAZ (unphosphorylated at Thr-232).
3D Structure:
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
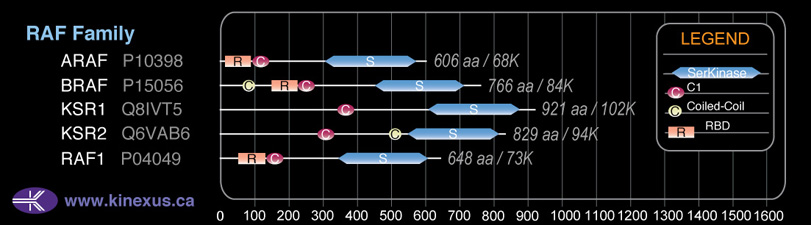
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S29-, S43-, S211, S220, S233+, S244, S247, S250, S252, S257, S259-, S267, S283, S285, S287, S289+, S291, S294, S295, S296-, S301-, S306, S338+, S339+, S357-, S359-, S471, S494+, S497+, S499+, S612, S619, S621+, S624, S642-.
Threonine phosphorylated:
T31, T49, T213, T234, T239, T242, T258, T260, T268, T269+, T303, T310, T324, T353, T362, T481+, T491+, T638, T640.
Tyrosine phosphorylated:
Y38, Y232, Y340+, Y341+, Y458.
Ubiquitinated:
K378, K493.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 33
33
1240
22
958
 5
5
192
14
104
 19
19
709
17
541
 16
16
609
106
949
 32
32
1192
21
660
 8
8
287
57
406
 16
16
580
37
615
 25
25
921
51
1182
 25
25
938
10
607
 9
9
338
88
242
 11
11
424
38
410
 18
18
655
163
665
 13
13
484
28
361
 8
8
290
12
180
 7
7
247
19
168
 5
5
197
14
149
 12
12
458
355
3102
 11
11
400
26
275
 24
24
895
74
794
 28
28
1047
79
668
 10
10
357
34
285
 12
12
434
37
397
 19
19
693
26
709
 12
12
431
26
319
 13
13
472
34
359
 22
22
807
72
772
 17
17
615
31
610
 12
12
442
26
293
 16
16
612
26
481
 1.5
1.5
55
14
38
 35
35
1295
18
681
 100
100
3717
26
6125
 14
14
515
67
721
 27
27
1008
57
789
 9
9
319
35
335
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.8
99.8
100
100 94.9
94.9
94.9
100 -
-
-
- -
-
-
- 98.8
98.8
99.2
99 -
-
-
- 97.7
97.7
98.3
98 98.3
98.3
98.5
98 -
-
-
- -
-
-
- 94.6
94.6
97.7
95 85.3
85.3
90.7
88 75.2
75.2
84
80 80.5
80.5
87
- 37.5
37.5
52.9
- -
-
-
- 36
36
51
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | YWHAZ - P63104 |
| 2 | HRAS - P01112 |
| 3 | YWHAB - P31946 |
| 4 | MAP2K1 - Q02750 |
| 5 | RAP1A - P62834 |
| 6 | HSP90AA2 - Q14568 |
| 7 | PEBP1 - P30086 |
| 8 | HSP90AA1 - P07900 |
| 9 | RRAS2 - P62070 |
| 10 | YWHAH - Q04917 |
| 11 | YWHAQ - P27348 |
| 12 | MAP2K2 - P36507 |
| 13 | PAK1 - Q13153 |
| 14 | BCL2 - P10415 |
| 15 | CDC25A - P30304 |
Regulation
Activation:
Activated by phosphorylation at Ser-338, Ser-339, Tyr-340, Tyr-341, Ser-471, Thr-481, Thr-491, Ser-494, Ser-497, Ser-499 and Ser-621. Phosphorylation of Ser-471 also induces interaction with MEK1. Phosphorylation of Ser-621 also induces interaction with 14-3-3-zeta.
Inhibition:
Phosphorylation at Ser-233 and Ser-289 inhibits interaction with H-Ras-1 and increases interaction with 14-3-3 beta. Phosphorylation at Ser-259 inhibits phosphotransferase activity, reduces H-Ras-1 and PPP2CB binding, and induces binding of 14-3-3-beta and zeta. It appears to necessary for PKC-alpha activation of Raf1. Phosphorylation of Ser-43, Ser-296, Ser-301 and Ser-642 inhibits phosphotransferase activity and inhibits interaction with H-Ras-1. Phosphorylation of Tyr-341 inhibits interaction with Raf1.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| PKACa | P17612 | S43 | FGYQRRASDDGKLTD | - |
| PKCa | P17252 | S43 | FGYQRRASDDGKLTD | - |
| PKACa | P17612 | S233 | VSSQHRYSTPHAFTF | + |
| PKACa | P17612 | S259 | SQRQRSTSTPNVHMV | - |
| AKT3 | Q9Y243 | S259 | SQRQRSTSTPNVHMV | - |
| Akt1 | P31749 | S259 | SQRQRSTSTPNVHMV | - |
| RAF1 | P04049 | S259 | SQRQRSTSTPNVHMV | - |
| KSR | Q8IVT5 | T269 | NVHMVSTTLPVDSRM | + |
| ERK1 | P27361 | S289 | RSHSESASPSALSSS | + |
| ERK1 | P27361 | S296 | SPSALSSSPNNLSPT | - |
| ERK1 | P27361 | S301 | SSSPNNLSPTGWSQP | - |
| MEK1 | Q02750 | S338 | RPRGQRDSSYYWEIE | + |
| RAF1 | P04049 | S338 | RPRGQRDSSYYWEIE | + |
| SRC | P12931 | S338 | RPRGQRDSSYYWEIE | + |
| PAK1 | Q13153 | S338 | RPRGQRDSSYYWEIE | + |
| PAK3 | O75914 | S338 | RPRGQRDSSYYWEIE | + |
| PAK1 | Q13153 | S339 | PRGQRDSSYYWEIEA | + |
| SRC | P12931 | Y340 | RGQRDSSYYWEIEAS | + |
| SRC | P12931 | Y341 | GQRDSSYYWEIEASE | + |
| PKCa | P17252 | S497 | ATVKSRWSGSQQVEQ | + |
| PKCa | P17252 | S499 | VKSRWSGSQQVEQPT | + |
| LCK | P06239 | S499 | VKSRWSGSQQVEQPT | + |
| PKCa | P17252 | S619 | SLPKINRSASEPSLH | + |
| PKACa | P17612 | S621 | PKINRSASEPSLHRA | + |
| RAF1 | P04049 | S621 | PKINRSASEPSLHRA | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ADCY6 | O43306 | S732 | PKALQRLSRSIVRSR | |
| ADCY6 | O43306 | S734 | ALQRLSRSIVRSRAH | |
| ADCY6 | O43306 | S738 | LSRSIVRSRAHSTAV | |
| ADCY6 | O43306 | S742 | IVRSRAHSTAVGIFS | |
| Bad | Q92934 | S75 | EIRSRHSSYPAGTED | - |
| MEK1 (MAP2K1) | Q02750 | S218 | VSGQLIDSMANSFVG | + |
| MEK1 (MAP2K1) | Q02750 | S222 | LIDSMANSFVGTRSY | + |
| MYPT1 | O14974 | T696 | ARQSRRSTQGVTLTD | |
| Raf1 | P04049 | S259 | SQRQRSTSTPNVHMV | - |
| Raf1 | P04049 | S338 | RPRGQRDSSYYWEIE | + |
| Raf1 | P04049 | S621 | PKINRSASEPSLHRA | + |
| TNNT2 | P45379 | T212 | RKSGKRQTEREKKKK |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
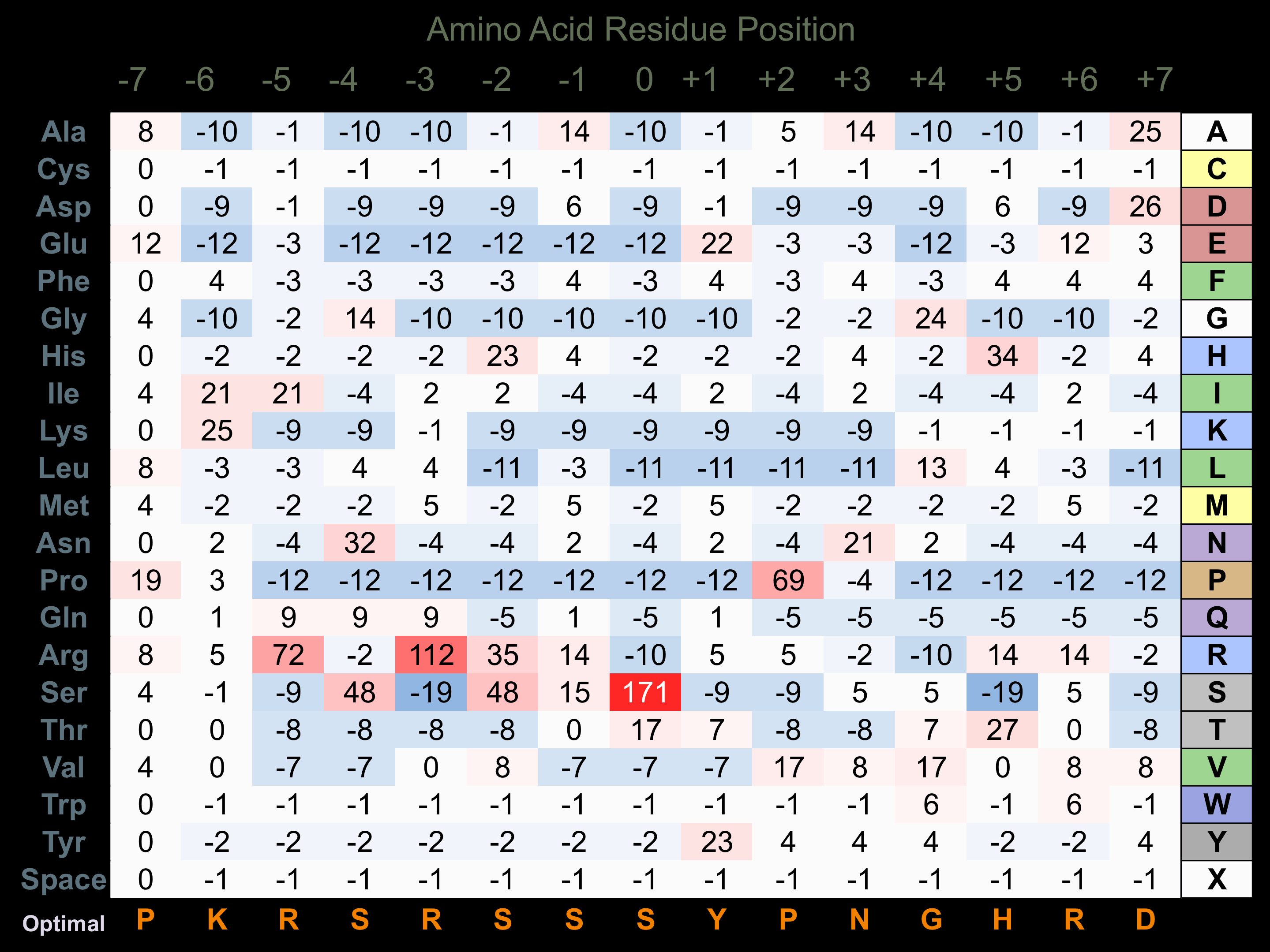
Matrix Type:
Experimentally derived from alignment of 19 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, genetic disorders
Specific Diseases (Non-cancerous):
Noonan syndrome 5; Leopard syndrome; Leopard syndrome 2; Noonan syndrome; Cardiofaciocutaneous (CFC) syndrome; Encephalitis; Dengue disease; St. Louis encephalitis; Pulmonic stenosis; West Nile fever; Murray valley encephalitis; Kyasanur forest disease; Noonan syndrome 1; Leopard syndrome 1; Cardiomyopathy, dilated, 1w; Raf1-related noonan syndrome; Raf1-related leopard syndrome
Comments:
Raf1 is linked to the rare Cardiofaciocutaneous Syndrome, which can affect the heart (cardio-), face (facio-), skin, and hair (cutaneous). Symptoms can include mild to severe cognitive disability, and developmental delay. Encephalitis is characterized by inflammation in the brain with symptoms ranging from flu-like to headache, fever, and vomiting. St. Louis Encephalitis is typically spread through a virus, is a rare neurological disorder, and is characterized by fever, headache, nausea, vomiting, and fatigue. Pulmonic stenosis links to congenital heart disease and is characterized by obstructed flow of blood from the right ventricle to the pulmonary artery. Kyasanur Forest Disease (KFD) is a rare infectious disease characterized by fever, headache, neck stiffness, muscle pain, cough, dehydration, and issue of bleeding. Cardiomyopathy, Dilated, 1w is a rare disorder characterized by stillbirth, and autosomal recessive x-linked dominant inheritance.
Specific Cancer Types:
Pilocytic astrocytomas (PA); Lung adenomas
Comments:
RAF1 appears to be an oncoprotein (OP). Cancer-related mutations in human tumours point to a gain of function of the protein kinase. The active form of the protein kinase normally acts to promote tumour cell proliferation. Overactive Raf-1 kinase phosphotransferase activity due to different mutations has been implicated in the pathogenesis of both Noonan and Leopard syndromes. >Noonan syndrome 5 (NS5) is a rare genetic disease that may manifest as cancer and is characterized by the abnormal development of multiple body parts resulting in distinctive facial construction, broad or webbed neck, short stature, bleeding issues, congenital heart anomalies, skeletal malformations, and developmental delay. In NS5, some common mutations observed include R256S, S259F, T260R, D486G, T491R, and S612T. The S257L or L613V mutations in NS5 have greater ERK1/2 (MAPK3/1) activation due to increased kinase phosphotransferase activity of Raf1. Its phosphotransferase activity can be increased, leading to enhanced ERK1/2 activation through a P261A, or P261L, or P261S, or a V263A mutation. In some cases of NS5, Raf1 kinase phosphotransferase activity can be decreased with a D486N or T491I mutation. >Leopard Syndrome is a rare yet very distinctive disorder where victims are marked with dark spots on their skin. They also suffer from abnormalities in the heart and genitalia, widely spaced eyes (ocular hypertelorism), pulmonary stenosis, short stature, and sensorineural deafness. S257L and L613V mutations are associated with Leopard syndrome 2, and these are also accompanied by increased downstream activation of ERK1/2. Many of the mutations in the Raf1 in both Noonan and Leopard syndromes are located in a motif flanking the S259 residue in the CR2 domain of the protein, which is critical for Raf-1 autoinhibition through 14-3-3 binding to phosphorylated S259. The S257L substitution mutation is also associated with other cancers. >Pilocytic Astrocytoma is a rare, benign, and slow growing tumour that develops in the brain or spinal cord. Inhibition of Raf1 phosphotransferase activity by the protein phosphatase PPP5C is impaired (no binding) with S338D and S339E mutations when these occur in conjunction with phosphomimetic Y340D and Y341D mutations in Raf1. This results in a constitutively active kinase, and extensive phosphorylation of S338. The T491D and T494D can increase kinase phosphotransferase activity but the kinase is still sensitive to PPP5C. Methylation of Raf1 can be inhibited with a R563K mutation, leading to increased stability and catalytic activity.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +132, p<0.0001); Brain glioblastomas (%CFC= -67, p<0.033); and Prostate cancer - primary (%CFC= +45, p<0.0001). The COSMIC website notes an up-regulated expression score for RAF1 in diverse human cancers of 560, which is 1.2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 401 for this protein kinase in human cancers was 6.7-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25725 diverse cancer specimens. This rate is very similar (+ 10% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.44 % in 1152 large intestine cancers tested; 0.33 % in 805 skin cancers tested; 0.31 % in 602 endometrium cancers tested; 0.31 % in 589 stomach cancers tested; 0.12 % in 1270 liver cancers tested; 0.1 % in 1941 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: S257L (13); P261R (4).
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.

