Nomenclature
Short Name:
p70S6K
Full Name:
Ribosomal protein S6 kinase 1
Alias:
- EC 2.7.11.1
- KS6B1
- S6K
- S6K1
- STK14A
- p70-alpha
- p70(S6K)-alpha
- Ribosomal protein S6 kinase I
- RPS6KB1
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
RSK
SubFamily:
p70
Specific Links
Structure
Mol. Mass (Da):
59,140
# Amino Acids:
525
# mRNA Isoforms:
5
mRNA Isoforms:
59,140 Da (525 AA; P23443); 56,593 Da (502 AA; P23443-4); 56,189 Da (502 AA; P23443-2); 52,997 Da (472 AA; P23443-3); 51,016 Da (451 AA; P23443-5)
4D Structure:
Interacts with PPP1R9A/neurabin-1 By similarity. Interacts with RPTOR. Interacts with IRS1. Interacts with EIF3B and EIF3C. Interacts with POLDIP3.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
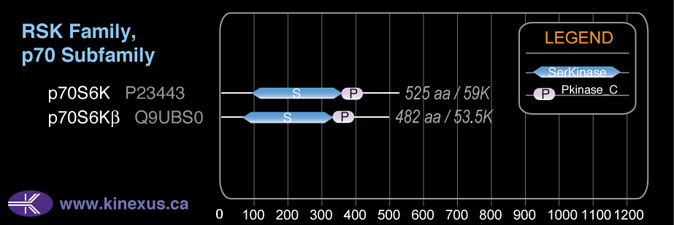
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K304, K516.
Serine phosphorylated:
S40+, S53, S243, S375, S380, S394+, S403, S417, S421, S427+, S434+, S441+, S447+, S452.
Threonine phosphorylated:
T248+, T250+, T252+, T256+, T390, T412+, T444+, T470.
Tyrosine phosphorylated:
Y62, Y413.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 29
29
975
35
997
 1
1
32
24
28
 5
5
157
16
196
 13
13
443
138
968
 23
23
785
41
687
 2
2
76
90
96
 9
9
302
55
461
 34
34
1147
65
2480
 10
10
331
23
356
 3
3
103
143
102
 2
2
70
52
93
 13
13
446
239
562
 3
3
104
38
162
 1.3
1.3
42
23
40
 2
2
76
32
113
 2
2
64
22
48
 1.4
1.4
48
390
173
 3
3
86
32
110
 3
3
110
140
124
 22
22
737
143
731
 3
3
86
47
136
 3
3
101
48
146
 4
4
120
26
116
 2
2
70
34
109
 3
3
101
47
156
 21
21
695
94
1125
 3
3
84
47
95
 4
4
122
31
217
 3
3
100
34
143
 2
2
72
42
71
 29
29
969
24
911
 100
100
3344
41
7153
 28
28
948
118
1322
 25
25
825
83
710
 4
4
126
48
166
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 85.9
85.9
85.9
100 94.3
94.3
95.6
- -
-
-
100 -
-
-
99 99.8
99.8
100
100 -
-
-
- 99.2
99.2
99.4
99 99.6
99.6
99.6
99 -
-
-
- 90.3
90.3
90.9
- 31.9
31.9
46.7
97 36.9
36.9
52.8
92.5 32.3
32.3
46.6
86 -
-
-
- 20.9
20.9
31.4
73 54.7
54.7
68
- 31.3
31.3
43.6
68 59
59
70.9
- -
-
-
- -
-
-
- -
-
-
49 36.4
36.4
55
49 -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | RPTOR - Q8N122 |
| 2 | DOCK5 - Q9H7D0 |
| 3 | MAPT - P10636 |
| 4 | NCBP1 - Q09161 |
| 5 | PDPK2 - Q6A1A2 |
| 6 | PDPK1 - O15530 |
| 7 | AKT1 - P31749 |
| 8 | RPS6 - P62753 |
| 9 | EEF2K - O00418 |
| 10 | EIF3B - P55884 |
| 11 | PPP1R9B - Q96SB3 |
| 12 | ACTA1 - P68133 |
| 13 | NEK6 - Q9HC98 |
| 14 | CDC2 - P06493 |
| 15 | PDK1 - Q15118 |
Regulation
Activation:
Phosphorylation at Thr-252, Ser-394, Thr-412, Ser-427, Ser-434, Ser-441, Thr-444 and Ser-447 increases phosphotransferase activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| CK2a1 | P68400 | S40 | DQPEDAGSEDELEEG | + |
| SRC | P12931 | Y62 | DHGGVGPYELGMEHC | |
| SRC | P12931 | T250 | SIHDGTVTHTFCGTI | + |
| PDK1 | O15530 | T252 | HDGTVTHTFCGTIEY | + |
| PIK3CD | O00329 | T252 | HDGTVTHTFCGTIEY | + |
| SRC | P12931 | S375 | PFKPLLQSEEDVSQF | |
| SRC | P12931 | S380 | LQSEEDVSQFDSKFT | |
| CDK1 | P06493 | S394 | TRQTPVDSPDDSTLS | + |
| FRAP1 | P42345 | S394 | TRQTPVDSPDDSTLS | + |
| FRAP1 | P42345 | T412 | NQVFLGFTYVAPSVL | + |
| p70S6K | P23443 | T412 | NQVFLGFTYVAPSVL | + |
| PDK1 | O15530 | T412 | NQVFLGFTYVAPSVL | + |
| PIK3CD | O00329 | T412 | NQVFLGFTYVAPSVL | + |
| NEK6 | Q9HC98 | T412 | NQVFLGFTYVAPSVL | + |
| JNK1 | P45983 | S434 | SFEPKIRSPRRFIGS | + |
| ERK2 | P28482 | S434 | SFEPKIRSPRRFIGS | + |
| CDK5 | Q00535 | S434 | SFEPKIRSPRRFIGS | + |
| JNK2 | P45984 | S434 | SFEPKIRSPRRFIGS | + |
| CDK1 | P06493 | S434 | SFEPKIRSPRRFIGS | + |
| FRAP1 | P42345 | S434 | SFEPKIRSPRRFIGS | + |
| ERK1 | P27361 | S434 | SFEPKIRSPRRFIGS | + |
| CDK1 | P06493 | S441 | SPRRFIGSPRTPVSP | + |
| ERK2 | P28482 | T444 | RFIGSPRTPVSPVKF | + |
| CDK1 | P06493 | T444 | RFIGSPRTPVSPVKF | + |
| ERK1 | P27361 | T444 | RFIGSPRTPVSPVKF | + |
| CDK5 | Q00535 | S447 | GSPRTPVSPVKFSPG | + |
| ERK2 | P28482 | S447 | GSPRTPVSPVKFSPG | + |
| CDK1 | P06493 | S447 | GSPRTPVSPVKFSPG | + |
| ERK1 | P27361 | S447 | GSPRTPVSPVKFSPG | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| BACH2 | Q9BYV9 | S521 | ETRTRTSSSCSSYSY | |
| Bad | Q92934 | S99 | PFRGRSRSAPPNLWA | - |
| CCT2 | P78371 | S260 | GSRVRVDSTAKVAEI | |
| CREM | Q03060-2 | S71 | EILSRRPSYRKILNE | |
| eEF2K | O00418 | S366 | SPRVRTLSGSRPPLL | - |
| eIF4B | P23588 | S422 | RERSRTGSESSQTGT | |
| ERa (ESR1) | P03372 | S167 | GGRERLASTNDKGSM | ? |
| FRAP1 (mTOR) | P42345 | S2448 | RSRTRTDSYSAGQSV | + |
| FRAP1 (mTOR) | P42345 | T2446 | NKRSRTRTDSYSAGQ | - |
| GluR1 | P42261 | T858 | NEAIRTSTLPRNSGA | |
| GSK3b | P49841 | S9 | SGRPRTTSFAESCKP | - |
| IRS1 | P35568 | S1101 | GCRRRHSSETFSSTP | - |
| IRS1 | P35568 | S270 | EFRPRSKSQSSSNCS | |
| IRS1 | P35568 | S307 | TRRSRTESITATSPA | - |
| IRS1 | P35568 | S312 | TESITATSPASMVGG | - |
| IRS1 | P35568 | S527 | RFRKRTHSAGTSPTI | ? |
| IRS1 | P35568 | S616 | DDGYMPMSPGVAPVP | - |
| IRS1 | P35568 | S636 | SGDYMPMSPKSVSAP | - |
| IRS1 | P35568 | S639 | YMPMSPKSVSAPQQI | - |
| LKB1 (STK11) | Q15831 | S428 | SSKIRRLSACKQQ__ | + |
| Mad1 (MXD1) | Q05195 | S145 | IERIRMDSIGSTVSS | - |
| NDRG2 | Q9UN36 | S332 | LSRSRTASLTSAASV | |
| NDRG2 | Q9UN36 | S350 | RSRSRTLSQSSESGT | |
| p63 | Q9H3D4 | S560 | LARLGCSSCLDYFTT | - |
| p70S6K (RPS6KB1) | P23443 | T412 | NQVFLGFTYVAPSVL | + |
| PDCD4 | Q53EL6 | S67 | KRRLRKNSSRDSGRG | |
| PFKFB2 | O60825 | S466 | PVRMRRNSFTPLSSS | |
| PFKFB2 | O60825 | S483 | IRRPRNYSVGSRPLK | |
| POLDIP3 | Q9BY77 | S383 | ELPRRVNSASSSNPP | |
| POLDIP3 | Q9BY77 | S385 | PRRVNSASSSNPPAE | |
| RSK1 (RPS6KA1) | Q15418 | S380 | HQLFRGFSFVATGLM | + |
| RSK2 (RPS6KA3) | P51812 | S386 | HQLFRGFSFVAITSD | + |
| S6 | P62753 | S235 | IAKRRRLSSLRASTS | + |
| S6 | P62753 | S236 | AKRRRLSSLRASTSK | + |
| S6 | P62753 | S240 | RLSSLRASTSKSESS | + |
| S6 | P62753 | S244 | LRASTSKSESSQK__ | + |
| S6 | P62753 | S247 | STSKSESSQK_____ | + |
| Tau iso8 | P10636-8 | S214 | GSRSRTPSLPTPPTR | |
| Tau iso8 | P10636-8 | S262 | NVKSKIGSTENLKHQ | |
| Tau iso8 | P10636-8 | T212 | TPGSRSRTPSLPTPP | |
| CAD | P27708 | S1859 | PPRIHRASDPGLPAE | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
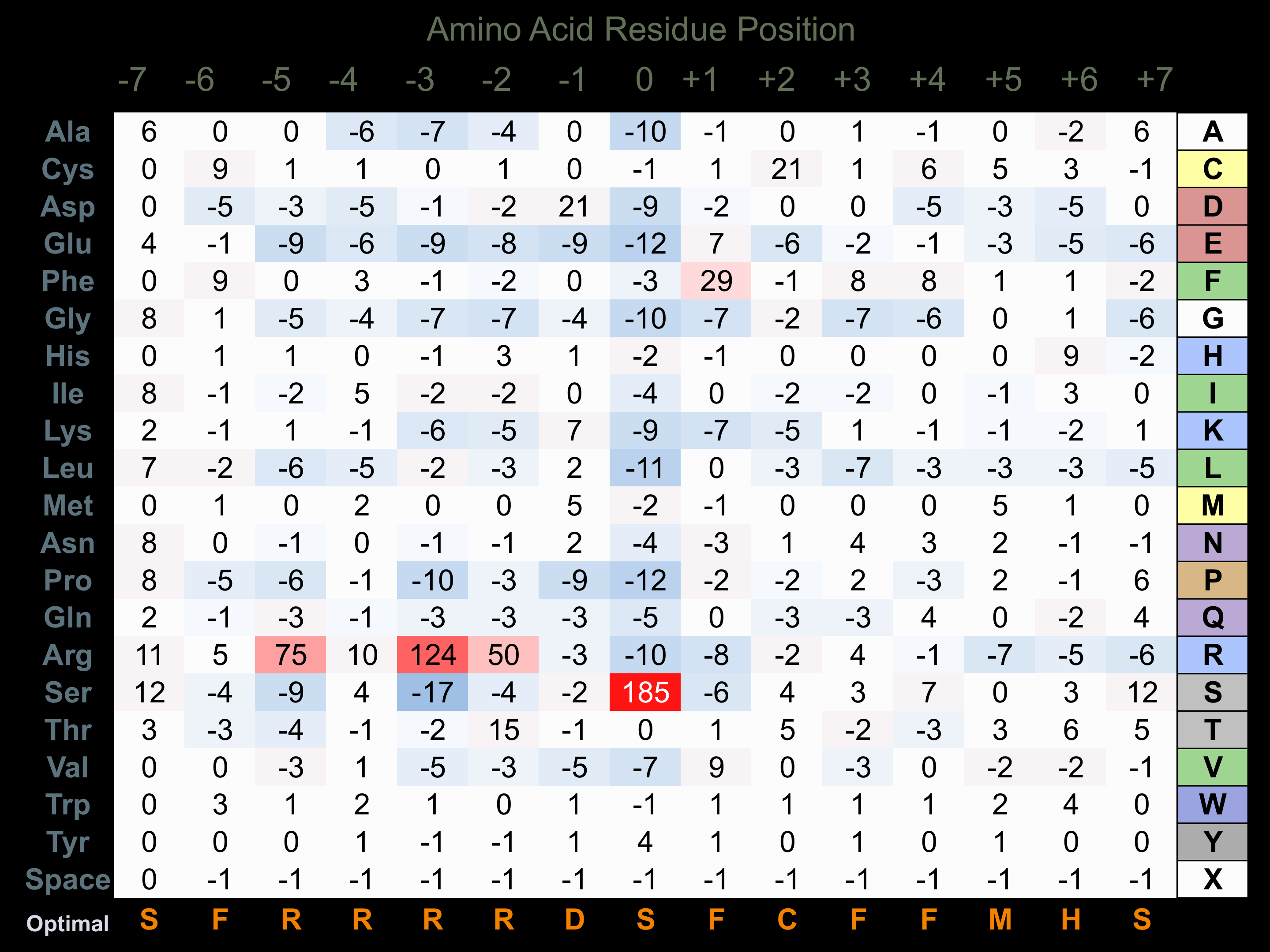
Matrix Type:
Experimentally derived from alignment of 58 known protein substrate phosphosites and 62 peptides phosphorylated by recombinant p70S6K in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Genetic disorders
Specific Diseases (Non-cancerous):
Tuberous sclerosis; Tuberous sclerosis complex
Comments:
Tuberous sclerosis is a rare genetic disease characterized by the growth of benign tumours (non-cancerous) in the brain and other organs. Symptoms are variable dependent upon location of tumour growth. Common symptoms include light-coloured patches of skin, areas of thickened skin, and tumour growth under the nails. This disease is inherited in an autosomal dominant manner. Known mutations in p70S6K are associated with a gain-of-function phenotype through the facilitation of phosphorylation by the upstream kinase PDPK1. These mutations are associated with tuberous sclerosis, potentially through the over-stimulation of cell growth and proliferation leading to aberrant proliferation of cells.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= -45, p<0.059); Cervical cancer (%CFC= +116, p<0.006); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +602, p<0.0001); Ovary adenocarcinomas (%CFC= +166, p<0.031); Pituitary adenomas (ACTH-secreting) (%CFC= -45); Prostate cancer - primary (%CFC= -87, p<0.0001); and Uterine leiomyomas from fibroids (%CFC= +66, p<0.035). The COSMIC website notes an up-regulated expression score for p70S6K in diverse human cancers of 766, which is 1.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 256 for this protein kinase in human cancers was 4.3-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 25555 diverse cancer specimens. This rate is only -35 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.2 % in 1119 large intestine cancers tested; 0.13 % in 1942 lung cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,718 cancer specimens
Comments:
Only 2 deletions, 1 insertion, and no complex mutations are noted on the COSMIC website.

