Nomenclature
Short Name:
p70S6Kb
Full Name:
Ribosomal protein S6 kinase 2
Alias:
- 14 beta
- EC 2.7.11.1
- RPS6KB2
- S6 kinase-related kinase
- S6K2
- S6K-beta 2; SRK; STK14B
- KLS
- KS6B2
- p70-beta-1
- p70-beta-2
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
RSK
SubFamily:
p70
Specific Links
Structure
Mol. Mass (Da):
53455
# Amino Acids:
482
# mRNA Isoforms:
2
mRNA Isoforms:
53,455 Da (482 AA; Q9UBS0); 16,868 Da (154 AA; Q9UBS0-2)
4D Structure:
NA
1D Structure:
Subfamily Alignment
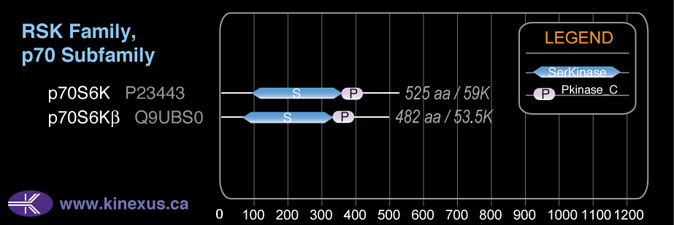
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K276.
Serine phosphorylated:
S15, S24, S219, S266, S356, S410+, S416, S417+, S423+, S473+.
Threonine phosphorylated:
T226+, T228+, T270.
Tyrosine phosphorylated:
Y45, Y96, Y136.
Ubiquitinated:
K86, K94.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 43
43
854
16
1176
 5
5
91
10
83
 7
7
138
15
165
 34
34
680
228
1745
 32
32
639
14
623
 21
21
418
44
989
 18
18
359
19
667
 52
52
1033
35
2514
 14
14
273
10
241
 8
8
165
59
210
 32
32
640
154
2045
 28
28
558
109
605
 6
6
119
26
115
 4
4
87
9
81
 5
5
97
11
110
 5
5
91
8
98
 9
9
186
116
120
 9
9
169
22
176
 29
29
579
232
1635
 30
30
588
56
601
 8
8
151
24
154
 8
8
151
26
127
 8
8
154
16
154
 32
32
642
190
1721
 9
9
176
24
152
 52
52
1024
41
2179
 5
5
97
29
105
 9
9
173
22
213
 6
6
121
22
124
 9
9
179
14
109
 47
47
932
18
634
 39
39
777
15
597
 7
7
145
50
411
 41
41
816
26
680
 100
100
1983
22
4251
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99
99
99.8
99 91.8
91.8
92.6
97 -
-
-
95 -
-
-
- 95
95
96.7
94 -
-
-
- 93
93
95
93 65.7
65.7
76
93 -
-
-
- 71
71
80.9
- 30.1
30.1
42.8
- 30.6
30.6
42.6
- 30.3
30.3
43.2
- -
-
-
- 21.1
21.1
28.3
- 56.9
56.9
70.8
- 30
30
43.1
- 61
61
74.5
- -
-
-
- -
-
-
- -
-
-
- 39.4
39.4
57.3
- -
-
-
50 -
-
-
43
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | BTK - Q06187 |
| 2 | SRC - P12931 |
| 3 | SYK - P43405 |
| 4 | PDPK2 - Q6A1A2 |
| 5 | PDPK1 - O15530 |
| 6 | RPS6 - P62753 |
| 7 | EEF2K - O00418 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| SRC | P12931 | Y45 | GLEPVGHYEEVELTE | |
| PDK1 | O15530 | T228 | HEGAVTHTFCGTIEY | + |
| PKCz | Q05513 | S473 | PPSGTKKSKRGRGRP | + |
| PKCi | P41743 | S473 | PPSGTKKSKRGRGRP | + |
| PKCa | P17252 | S473 | PPSGTKKSKRGRGRP | + |
| PKCb | P05771 | S473 | PPSGTKKSKRGRGRP | + |
| PKCg | P05129 | S473 | PPSGTKKSKRGRGRP | + |
| PKCe | Q02156 | S473 | PPSGTKKSKRGRGRP | + |
| PKCd | Q05655 | S473 | PPSGTKKSKRGRGRP | + |
| PKCb | P05771 | S473 | PPSGTKKSKRGRGRP | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
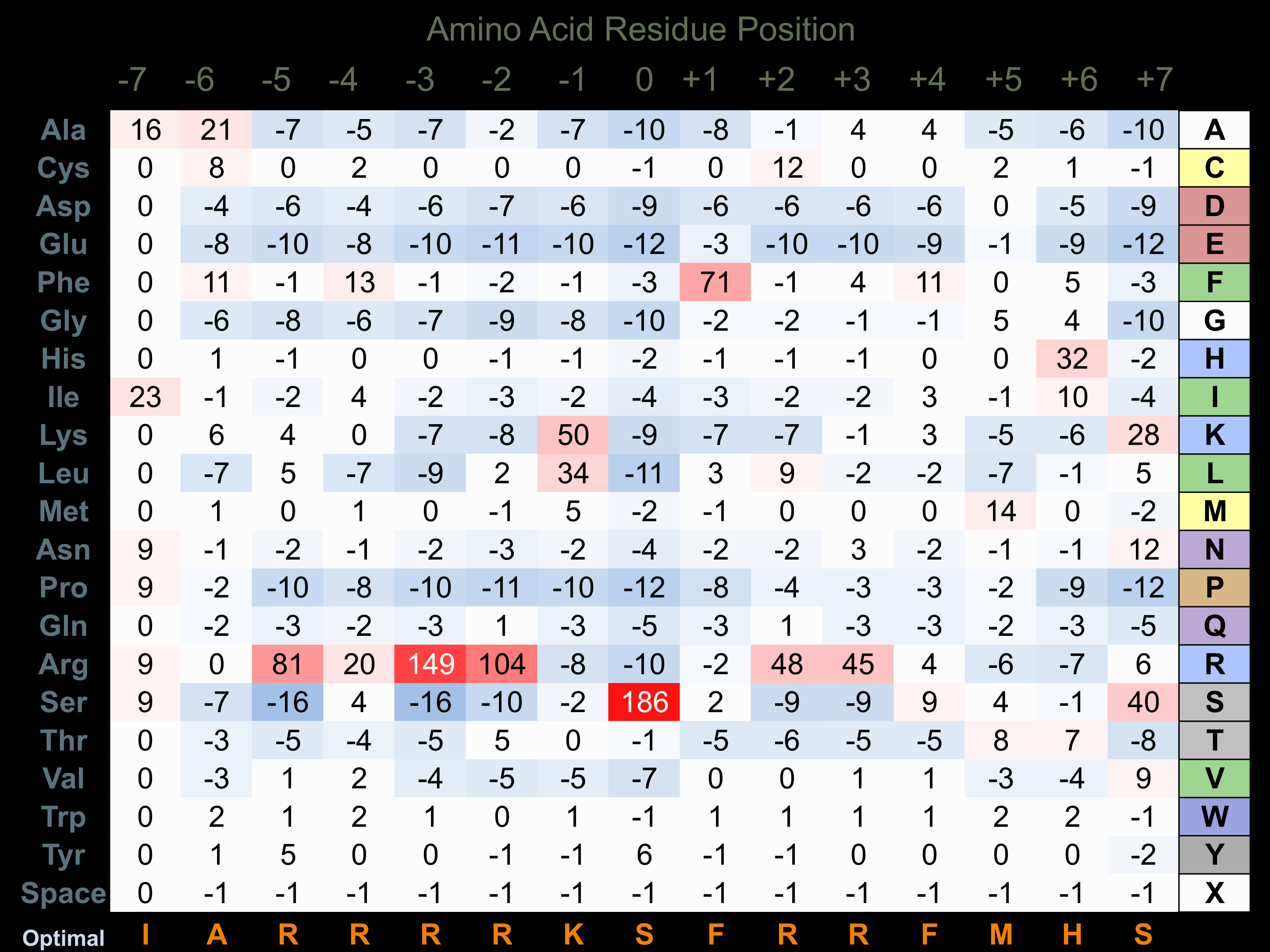
Matrix Type:
Experimentally derived from alignment of 9 known protein substrate phosphosites and 15 peptides phosphorylated by recombinant p70S6Kb in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Staurosporine | IC50 = 5 nM | 5279 | ||
| CHEMBL435809 | IC50 = 100 nM | 44352842 | 435809 | |
| Gö6976 | IC50 > 150 nM | 3501 | 302449 | 22037377 |
| K-252a; Nocardiopsis sp. | IC50 > 250 nM | 3813 | 281948 | 22037377 |
| SB218078 | IC50 > 250 nM | 447446 | 289422 | 22037377 |
| Bisindolylmaleimide I | IC50 > 1 µM | 2396 | 7463 | 22037377 |
| CK7 | Ki > 1 µM | 447961 | 15027857 | |
| Gö6983 | IC50 > 1 µM | 3499 | 261491 | 22037377 |
| Ro-32-0432 | IC50 > 1 µM | 127757 | 26501 | 22037377 |
| JNK Inhibitor VIII | Ki = 3.4 µM | 11624601 | 210618 | 16759099 |
Disease Linkage
General Disease Association:
Endocrine, bone, and neuronal disorders
Specific Diseases (Non-cancerous):
Coffin-Lowry syndrome (CLS); Kleine-Levin syndrome; Landau-Kleffner syndrome (LKS)
Comments:
The rare syndrome Coffin-Lowry Syndrome (CLS) is characterized by cognitive impairment, short stature, head, face, and skeletal abnormalities (including upper jaw deformation and curvature of the spine), broad nose, extensive brow, eyelid folds that are slanting downwards, widely-spaced eyes, and thick eyebrows. CLS can lead to issues eating, respiratory issues, cognitive impairment, developmental delay, impaired hearing, strange gait, stimulus-induced drop episodes, and heart or kidney issues. Kleine-Levin Syndrome can be characterized by quick onset of flu-like symptoms, increased food consumption, irritability, disorientation, hallucinations, childishness, and uninhibited sex drive. Kleine-Levin Syndrome (LKS) has a relation to autism and aphasia. LKS is characterized by slow onset of aphasia (inability to understand or articulate language), and periodically characterized by developmental anomalies, impaired hearing, learning disability, attention deficit disorder, and cognitive impairment.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for p70S6Kb in diverse human cancers of 563, which is 1.2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 21 for this protein kinase in human cancers was 0.4-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 25411 diverse cancer specimens. This rate is only -18 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.3 % in 1093 large intestine cancers tested.
Frequency of Mutated Sites:
None > 6 in 20,677 cancer specimens
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.

