Nomenclature
Short Name:
GSK3A
Full Name:
Glycogen synthase kinase-3 alpha
Alias:
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
GSK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
50,981
# Amino Acids:
483
# mRNA Isoforms:
1
mRNA Isoforms:
50,981 Da (483 AA; P49840)
4D Structure:
Monomer. Interacts with ARRB2
3D Structure:
1D Structure:
Subfamily Alignment
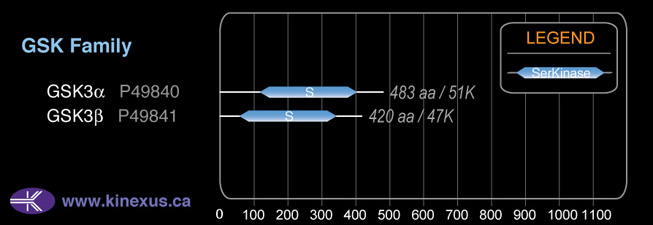
Domain Distribution:
| Start | End | Domain |
|---|
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S2, S7, S14, S20, S21-, S39, S41, S52, S63, S72, S77, S97, S266, S278+, S282-.
Threonine phosphorylated:
T19, T46, T338, T340, T480.
Tyrosine phosphorylated:
Y134, Y177, Y180, Y197, Y220, Y226, Y279+, Y284-, Y285-, Y351.
Ubiquitinated:
K268, K355, K360.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 69
69
907
32
1131
 15
15
198
15
182
 18
18
232
12
114
 35
35
465
91
560
 67
67
885
25
701
 5
5
63
74
40
 18
18
234
31
478
 62
62
820
40
984
 53
53
699
17
531
 16
16
206
83
193
 14
14
191
31
227
 57
57
756
184
668
 10
10
127
34
59
 16
16
206
12
190
 17
17
228
25
139
 13
13
166
15
133
 13
13
170
119
83
 19
19
254
21
203
 14
14
186
94
125
 46
46
605
109
581
 20
20
266
23
156
 13
13
177
27
140
 21
21
278
22
162
 19
19
249
21
175
 16
16
206
23
134
 49
49
650
57
670
 11
11
147
37
86
 18
18
234
21
163
 16
16
211
21
133
 77
77
1011
28
599
 24
24
314
24
243
 100
100
1321
36
2330
 6
6
82
72
209
 63
63
836
57
731
 12
12
163
35
180
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.6
99.6
99.8
100 99.8
99.8
99.8
100 -
-
-
96 -
-
-
98 85.5
85.5
86.3
96.5 -
-
-
- 93.9
93.9
95.5
95 97.1
97.1
97.7
97 -
-
-
- -
-
-
- 67.9
67.9
75
- 67.5
67.5
74.5
86 71.8
71.8
77.6
88.5 -
-
-
- 33.5
33.5
38
- 64.2
64.2
75.2
- 55.3
55.3
63.8
- -
-
-
- -
-
-
- 24.4
24.4
38.1
- -
-
-
67 52.8
52.8
63.8
67 40.8
40.8
55.5
59 -
-
-
70
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Phosphorylation at Thr-279 increases phosphotransferase activity and promotes nuclear localization.
Inhibition:
Phosphorylation at Ser-21 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| PKACa | P17612 | S21 | SGRARTSSFAEPGGG | - |
| Akt1 | P31749 | S21 | SGRARTSSFAEPGGG | - |
| PKCz | Q05513 | S21 | SGRARTSSFAEPGGG | - |
| RSK2 | P51812 | S21 | SGRARTSSFAEPGGG | - |
| PKCb | P05771 | S21 | SGRARTSSFAEPGGG | - |
| PKCd | Q05655 | S21 | SGRARTSSFAEPGGG | - |
| PKCb | P05771 | S21 | SGRARTSSFAEPGGG | - |
| PKCg | P05129 | S21 | SGRARTSSFAEPGGG | - |
| PKCh | P24723 | S21 | SGRARTSSFAEPGGG | - |
| MEK2 | P36507 | Y279 | RGEPNVSYICSRYYR | + |
| MEK1 | Q02750 | Y279 | RGEPNVSYICSRYYR | + |
| GSK3A | P49840 | Y279 | RGEPNVSYICSRYYR | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ACLY | P53396 | S451 | STSTPAPSRTASFSE | |
| ACLY | P53396 | T447 | NASGSTSTPAPSRTA | |
| ANKRD28 | O15084 | S1040 | INRYTNTSKTVSFEA | |
| Axin | O15169 | S486 | LRTPGRQSPGPGHRS | |
| Axin | O15169 | T481 | HVQRVLRTPGRQSPG | |
| Bcl-3 | P20749 | S394 | LSASPSSSPSQSPPR | |
| Bcl-3 | P20749 | S398 | PSSSPSQSPPRDPPG | |
| C-EBPa | P49715 | T226 | HLQPGHPTPPPTPVP | |
| C-EBPa | P49715 | T230 | GHPTPPPTPVPSPHP | |
| CdGAP | Q9ULL6 Q2M1Z3 | T790 | PPAPPPPTPLEESTP | |
| CREB1 | P16220 | S129 | QKRREILSRRPSYRK | + |
| CRMP2 (DPYSL2) | Q16555 | S518 | KTVTPASSAKTSPAK | |
| CTNNB1 | P35222 | S33 | QQQSYLDSGIHSGAT | - |
| CTNNB1 | P35222 | S37 | YLDSGIHSGATTTAP | ? |
| CTNNB1 | P35222 | T41 | GIHSGATTTAPSLSG | ? |
| Cyclin D1 (CCND1) | P24385 | T286 | EEVDLACTPTDVRDV | |
| Cyclin E1 (CCNE1) | P24864 | T395 | PLPSGLLTPPQSGKK | |
| eIF2B-e | Q13144 | S540 | MDSEEPDSRGGSPQM | - |
| GSK3a | P49840 | Y279 | RGEPNVSYICSRYYR | + |
| IRS2 | Q9Y4H2 | S487 | RPSSGSASASGSPSD | |
| MAP2 | P11137 | S136 | ETANLPPSPPPSPAS | |
| MAP2 | P11137 | T1616 | YSSRTPGTPGTPSYP | |
| MAP2 | P11137 | T1619 | RTPGTPGTPSYPRTP | |
| MITF | O75030 | S405 | QARAHGLSLIPSTGL | + |
| MKI67IP (NIFK) | Q9BYG3 | S230 | TPEKTVDSQGPTPVC | |
| MKI67IP (NIFK) | Q9BYG3 | T234 | TVDSQGPTPVCTPTF | ? |
| Myc | P01106 | T58 | KKFELLPTPPLSPSR | |
| NDRG1 | Q92597 | S342 | TSLDGTRSRSHTSEG | |
| NDRG1 | Q92597 | S352 | HTSEGTRSRSHTSEG | |
| NDRG1 | Q92597 | S362 | HTSEGTRSRSHTSEG | |
| Neurogenin 2 | Q9H2A3 | S234 | SCTLSPASPGSDVDY | |
| Neurogenin 2 | Q9H2A3 | S239 | SPYSCTLSPASPAGS | |
| PKAR2A | P13861 | S48 | REARAPASVLPAATP | |
| Polycystin 2 | Q13563 | S76 | AGAAASPSPPLSSCS | |
| PPP1R2 | P41236 | T72 | MKIDEPSTPYHSMIG | |
| PXN | P49023 | S126 | SFPNKQKSAEPSPTV | |
| Tau iso8 | P10636-8 | S262 | NVKSKIGSTENLKHQ | |
| Tau iso8 | P10636-8 | T231 | KKVAVVRTPPKSPSS | |
| TBX21 | Q9UL17 | S513 | PSSGDSSSPAGAPSP | ? |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
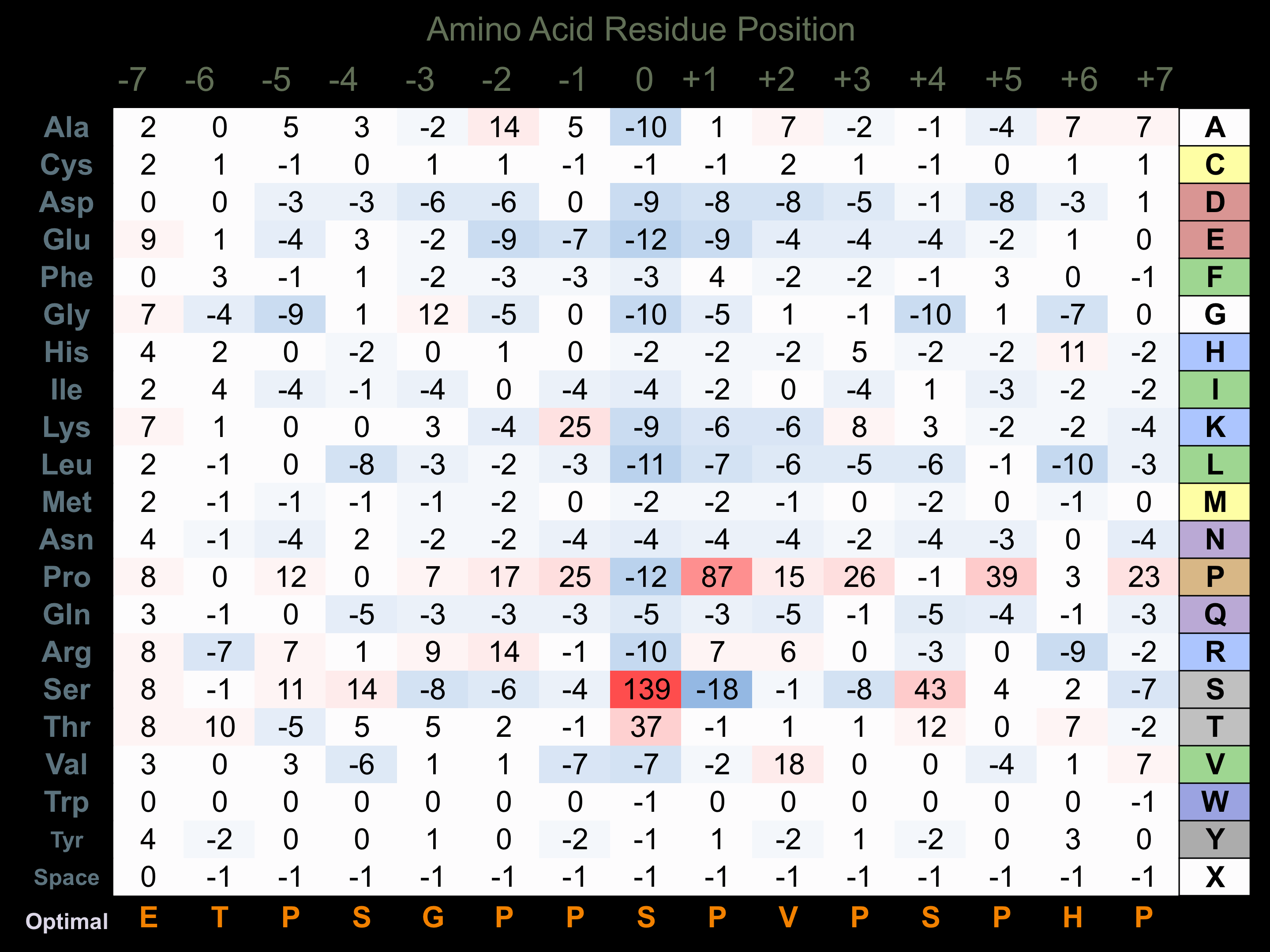
Matrix Type:
Experimentally derived from alignment of 59 known protein substrate phosphosites and 11 peptides phosphorylated by recombinant GSK3-alpha in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, neurological, and endocrine disorders
Specific Diseases (Non-cancerous):
Schizophrenia; Alzheimers disease (AD); Diabetes mellitus
Comments:
In Alzheimer’s Disease GSK3a is essential for maximal production of the aggregating proteins, amyloid-40 and -42.
Specific Cancer Types:
Sarcomas, Breast cancer
Comments:
GSK3 has been proposed as a tumour suppressor protein (TSP), since with loss of GSK3 there is an increase in WNT signalling, with a beta-catenin stabilization, leading to tumorigenesis. GSK3 has also been proposed to act as an oncoprotein (OP), as acute leukemia required GSK3 for cell proliferation. In most human cancers, it is up-regulated in expression about 1.6-times the rate observed for most protein kinases.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Cervical cancer (%CFC= +57, p<(0.0003); Ovary adenocarcinomas (%CFC= +88, p<0.0008); T-cell prolymphocytic leukemia (%CFC= +68, p<0.028); and Uterine leiomyosarcomas (%CFC= +52, p<0.021). The COSMIC website notes an up-regulated expression score for GSK3a in diverse human cancers of 725, which is 1.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 167 for this protein kinase in human cancers was 2.8-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 25371 diverse cancer specimens. This rate is only -16 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.41 % in 1270 large intestine cancers tested; 0.34 % in 603 endometrium cancers tested; 0.21 % in 589 stomach cancers tested; 0.15 % in 548 urinary tract cancers tested; 0.07 % in 864 skin cancers tested; 0.07 % in 1512 liver cancers tested; 0.06 % in 1276 kidney cancers tested; 0.05 % in 1956 lung cancers tested.
Frequency of Mutated Sites:
None > 5 in 20,654 cancer specimens
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.

